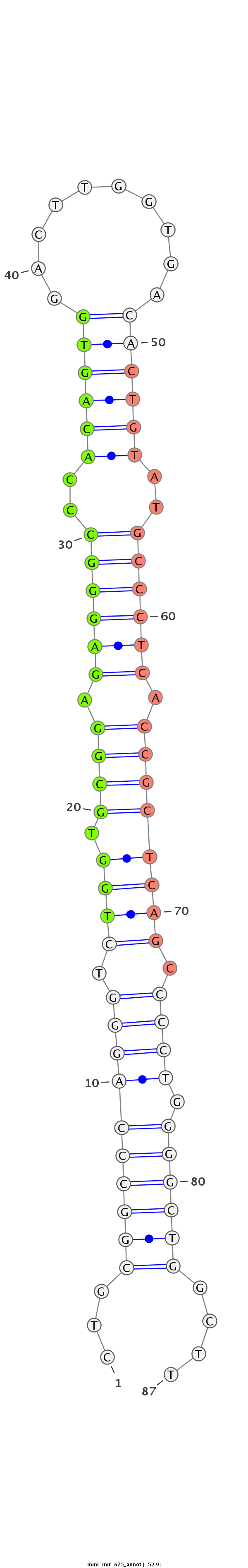
ID:mml-mir-675 |
Coordinate:chr14:1861015-1861087 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -52.9 | -52.9 | -52.9 |
 |
 |
 |
|
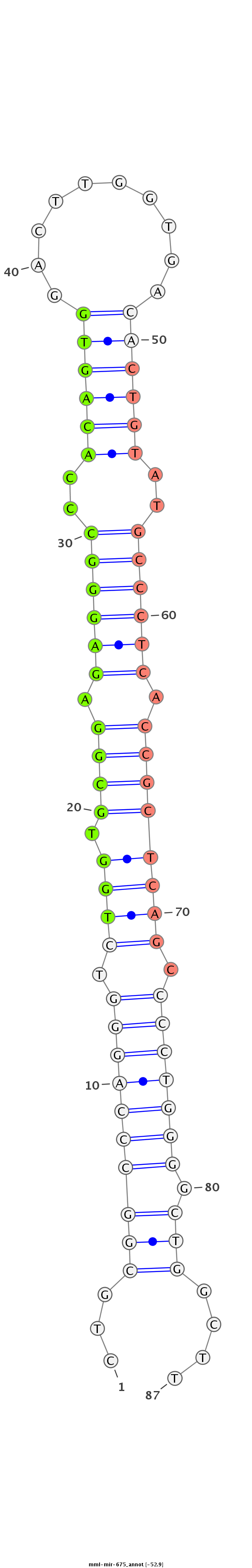
GAGGCTGCAGGGGCTCGGCCTGCGGGCGCCGTTCCCACGAGGCACTGCGGCCCAGGGTCTGGTGCGGAGAGGGCCCACAGTGGACTTGGTGACACTGTATGCCCTCACCGCTCAGCCCCTGGGGCTGGCTTGGCAGGCAGTACAGCATCCAGGGGAGTCAAGGGCATGGGGCG
********************************************...((((((((((.((((.((((.((((((..((((((..........))))))..)))))).)))))))).)))).))))))....****************************************** |
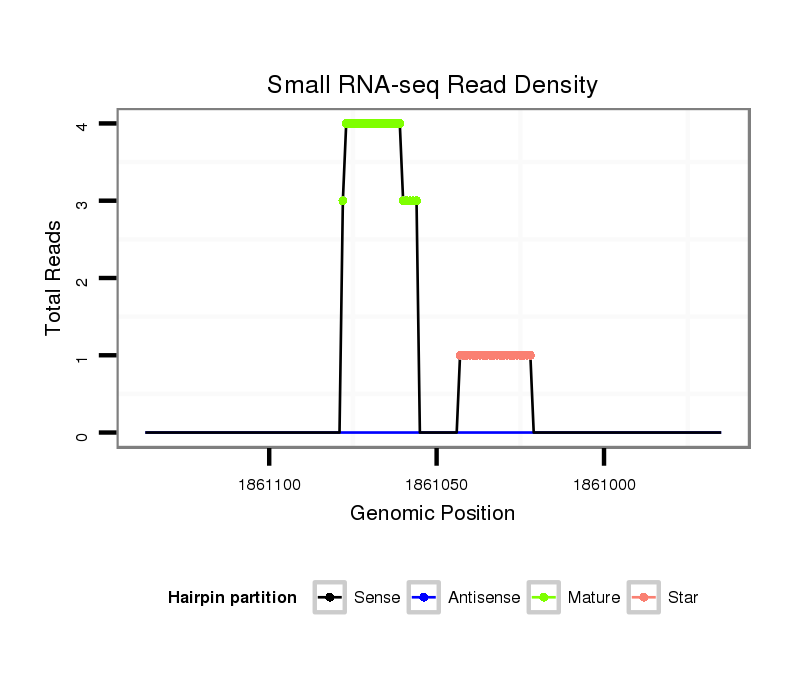
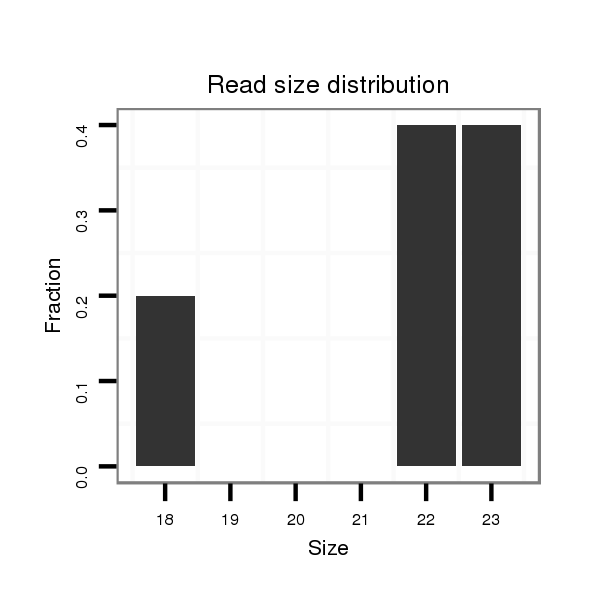
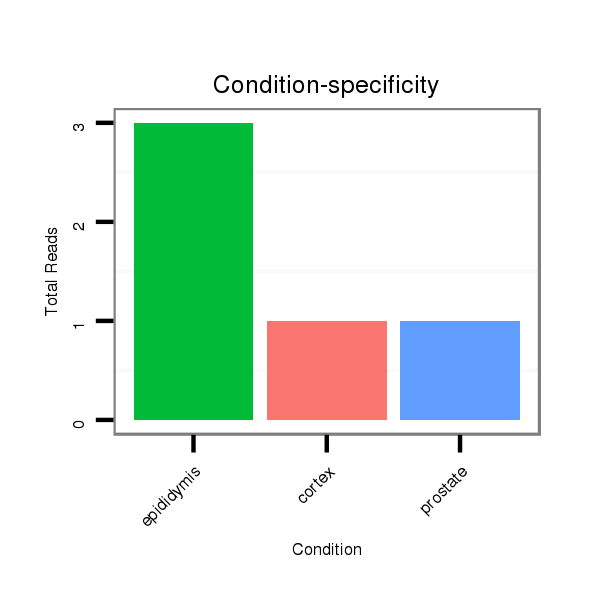
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116844 epididymis |
SRR116840 epididymis |
SRR116841 prostate |
SRR116843 cortex |
SRR116845 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................CTGTATGCCCTCACCGCTCAGCATC...................................................... | 25 | 2 | 1 | 8.00 | 8 | 7 | 0 | 0 | 0 | 1 |
| ...........................................................TGGTGCGGAGAGGGCCCACAGTG........................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 |
| ............................................................GGTGCGGAGAGGGCCCACAGTG........................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................TGGTGCGGAGAGGGCCCA................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................CTGTATGCCCTCACCGCTCAGC......................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................TTGTATGCCCTCACCGCTCAGCATC...................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................GTATGCCCTCACCGCTCAGCATC...................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................TGTATGCCCTCACCGCTCAGCATC...................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................TGGTGCGGAGAGGGCCCACAGTGT.......................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................TGTATTCCCTCACCGCTCAGCATC...................................................... | 24 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................CTGTATGCCCTCACCGCTCAGAAA....................................................... | 24 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
|
CTCCGACGTCCCCGAGCCGGACGCCCGCGGCAAGGGTGCTCCGTGACGCCGGGTCCCAGACCACGCCTCTCCCGGGTGTCACCTGAACCACTGTGACATACGGGAGTGGCGAGTCGGGGACCCCGACCGAACCGTCCGTCATGTCGTAGGTCCCCTCAGTTCCCGTACCCCGC
******************************************...((((((((((.((((.((((.((((((..((((((..........))))))..)))))).)))))))).)))).))))))....******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:09 PM