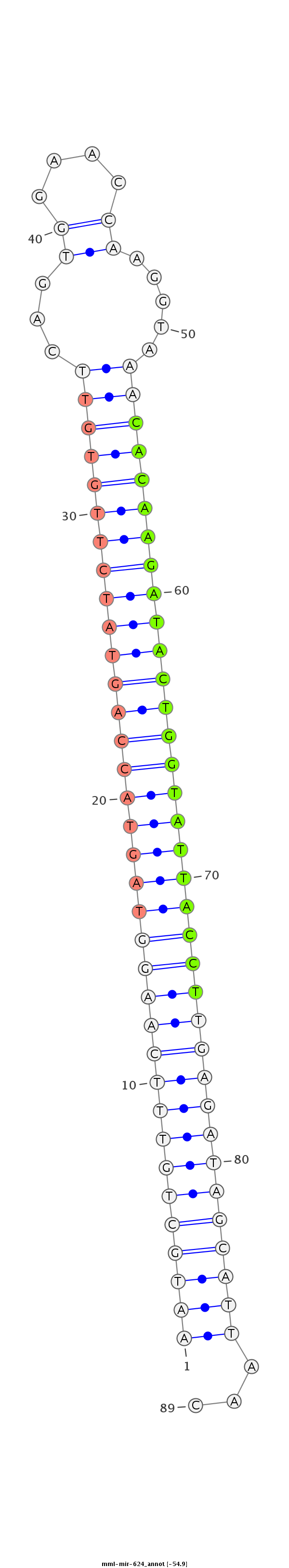
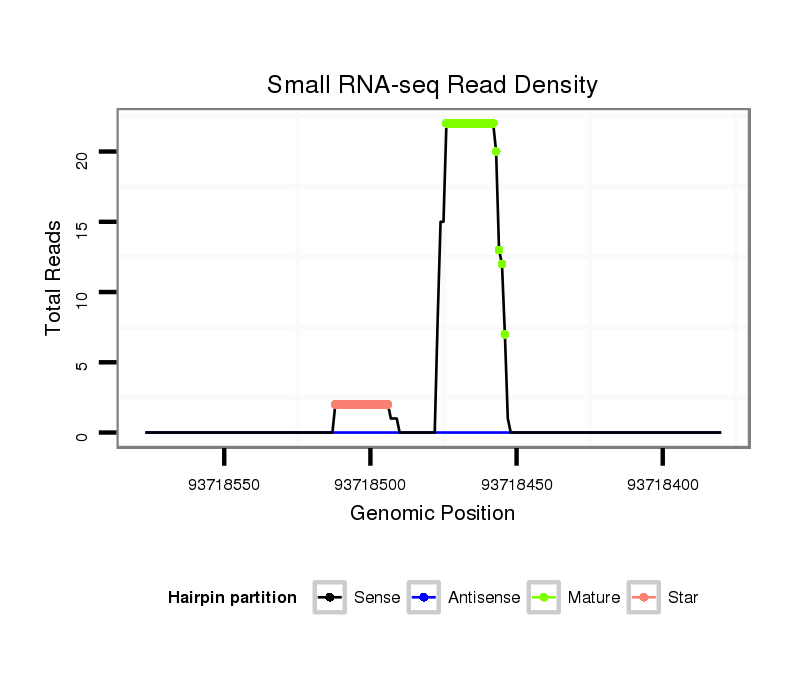
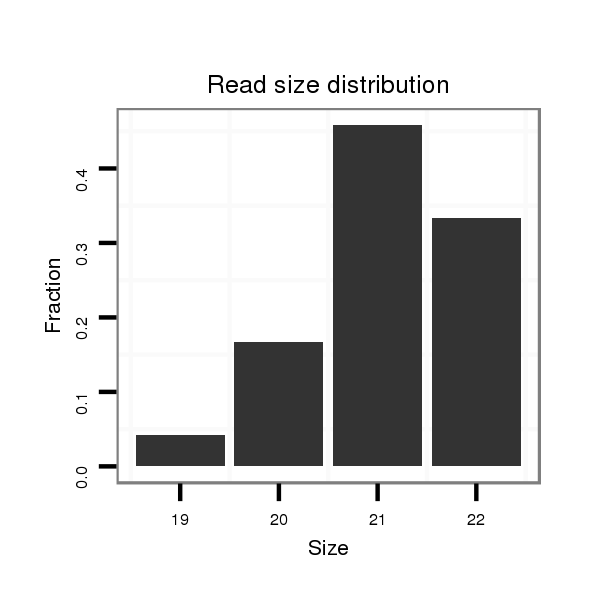
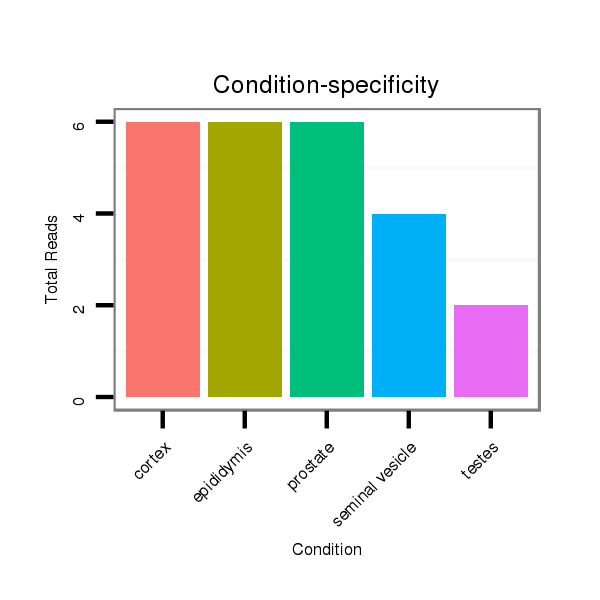
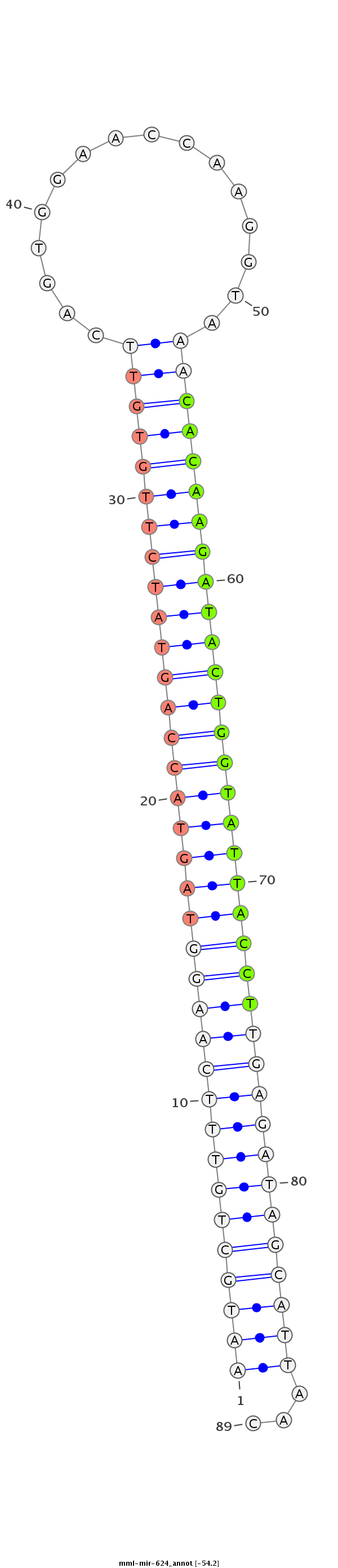
ID:mml-mir-624 |
Coordinate:chr7:93718430-93718527 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -54.2 | -54.2 |
 |
 |
|
TAATGAAAAAGAAAAAAGGTTTAAAAAGGATGTGCTTTTGACCAATATGTAATGCTGTTTCAAGGTAGTACCAGTATCTTGTGTTCAGTGGAACCAAGGTAAACACAAGATACTGGTATTACCTTGAGATAGCATTAACACCTAAGTGGTTTCAGTGAACAAGATTTAAAAGACTAGAATTAAAGCAATTATAAATGT
**************************************************(((((((((((((((((((((((((((((((((((...((....)).....)))))))))))))))))))))))))))))))))))...*********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116841 prostate |
SRR116843 cortex |
SRR116840 epididymis |
SRR116842 seminal vesicle |
SRR116839 testes |
SRR116845 epididymis |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................CACAAGATACTGGTATTACCT.......................................................................... | 21 | 0 | 1 | 6.00 | 6 | 1 | 2 | 1 | 1 | 1 | 0 | 0 |
| .....................................................................................................AACACAAGATACTGGTATTACC........................................................................... | 22 | 0 | 1 | 5.00 | 5 | 2 | 1 | 0 | 1 | 1 | 0 | 0 |
| ....................................................................................................AAACACAAGATACTGGTATTA............................................................................. | 21 | 0 | 1 | 5.00 | 5 | 1 | 2 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAACACAAGATACTGGTATT.............................................................................. | 20 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................AACACAAGATACTGGTATTACCATC........................................................................ | 25 | 2 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ....................................................................................................AAACACAAGATACTGGTATTAT............................................................................ | 22 | 1 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................AACACAAGATACTGGTATTA............................................................................. | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| .................................................................TAGTACTAGTATCTTGTGTTCAA.............................................................................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TAGTACCAGTATCTTGTGT.................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................TAGTACCAGTATCTTGTGTTCA............................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TAGTACCAGTATCTTTTGTTCA............................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AACACAAGATACTGGTATTACT........................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................AAACACAACATACTGGTATTA............................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAACACAAGATGCTGGTATTAC............................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAACACAAGATACTGGTATTAC............................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................CACAAGATACTGGTATTACCTT......................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................CACAGGATACTGGTATTACCT.......................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................CACAAGATACTGGTATTACCTTT........................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................GTAGTACCAGTATCTTGTGTTATC.............................................................................................................. | 24 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................AAACACAGGATACTGGTATTA............................................................................. | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
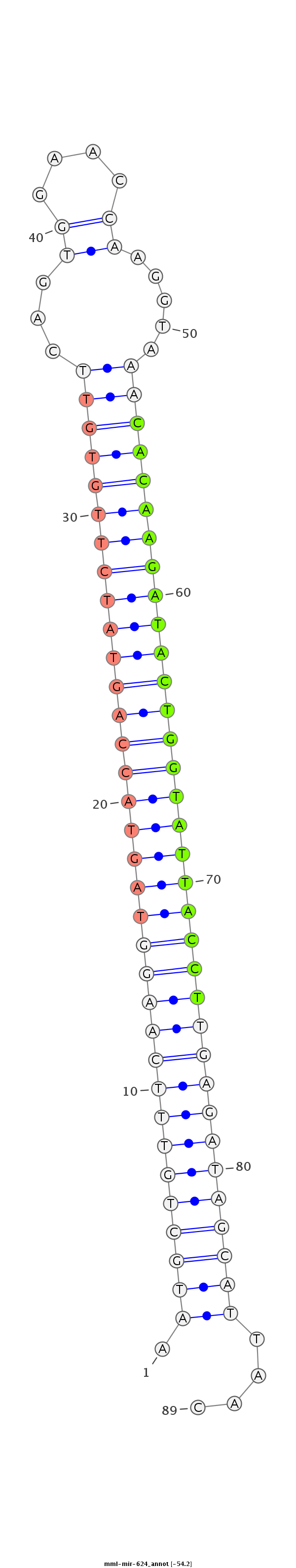
ATTACTTTTTCTTTTTTCCAAATTTTTCCTACACGAAAACTGGTTATACATTACGACAAAGTTCCATCATGGTCATAGAACACAAGTCACCTTGGTTCCATTTGTGTTCTATGACCATAATGGAACTCTATCGTAATTGTGGATTCACCAAAGTCACTTGTTCTAAATTTTCTGATCTTAATTTCGTTAATATTTACA
***********************************************************(((((((((((((((((((((((((((((((((((...((....)).....)))))))))))))))))))))))))))))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116844 epididymis |
|---|---|---|---|---|---|---|
| ..................................................................................................CTATTGTGTTCTATGACCATGATG............................................................................ | 24 | 3 | 2 | 0.50 | 1 | 1 |
Generated: 09/01/2015 at 12:27 PM