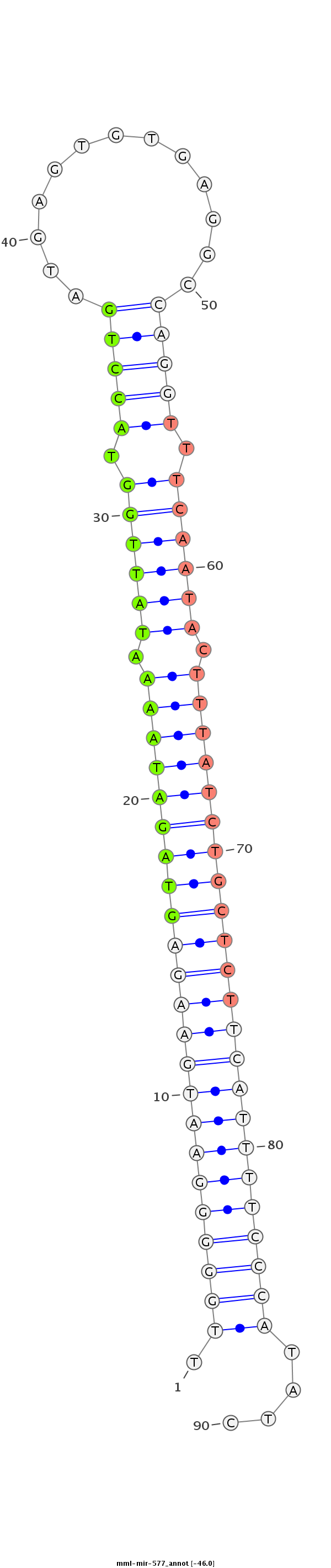
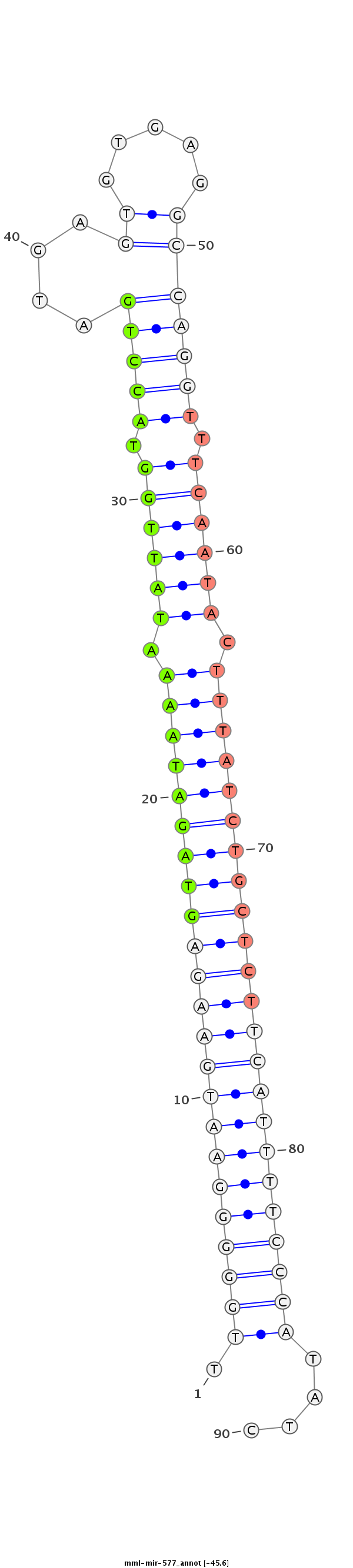
ID:mml-mir-577 |
Coordinate:chr5:108859184-108859279 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
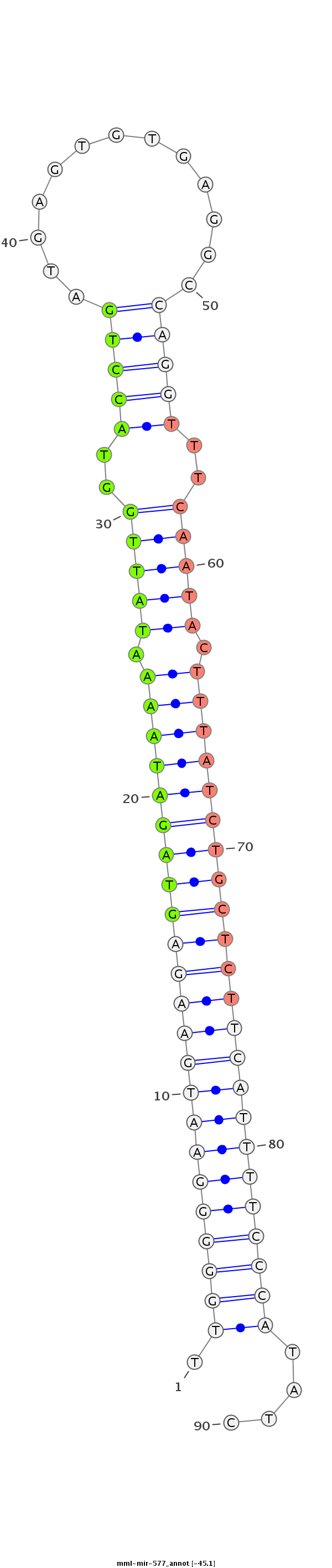
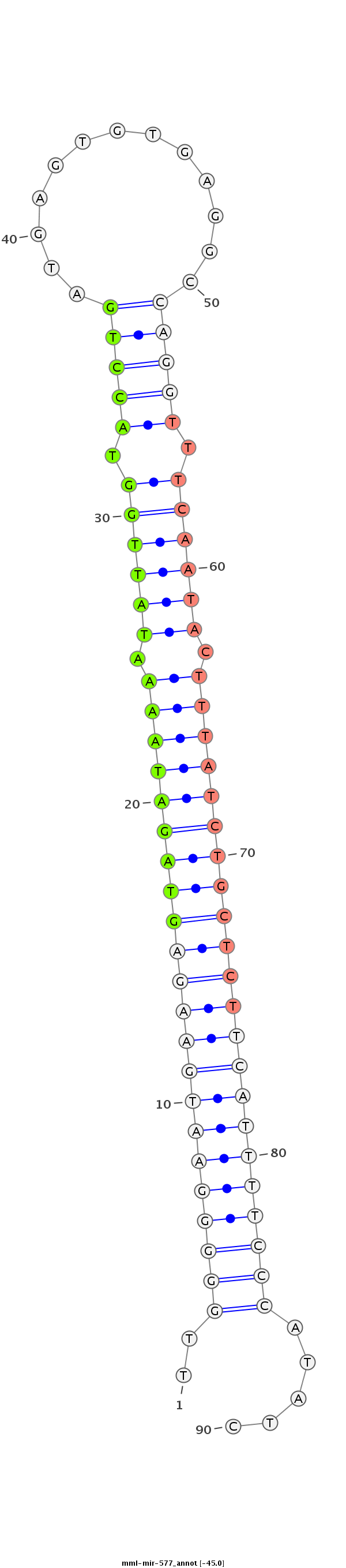
| -45.6 | -45.1 | -45.0 |
 |
 |
 |
|
ATGTGCTGGGGGGCGGAGGAAGTTTCAGGTACCCAATGGCAGGCTGCATTTGGGGGAATGAAGAGTAGATAAAATATTGGTACCTGATGAGTGTGAGGCCAGGTTTCAATACTTTATCTGCTCTTCATTTTCCCATATCTACTTACTTTAGGTATGGCCTCATTAAAAAATATAAACTCTCTGAAGACAGAACATT
*************************************************.(((((((((((((((((((((((.((((((.(((((.............))))).)))))).)))))))))))))))))))))))....********************************************************* |
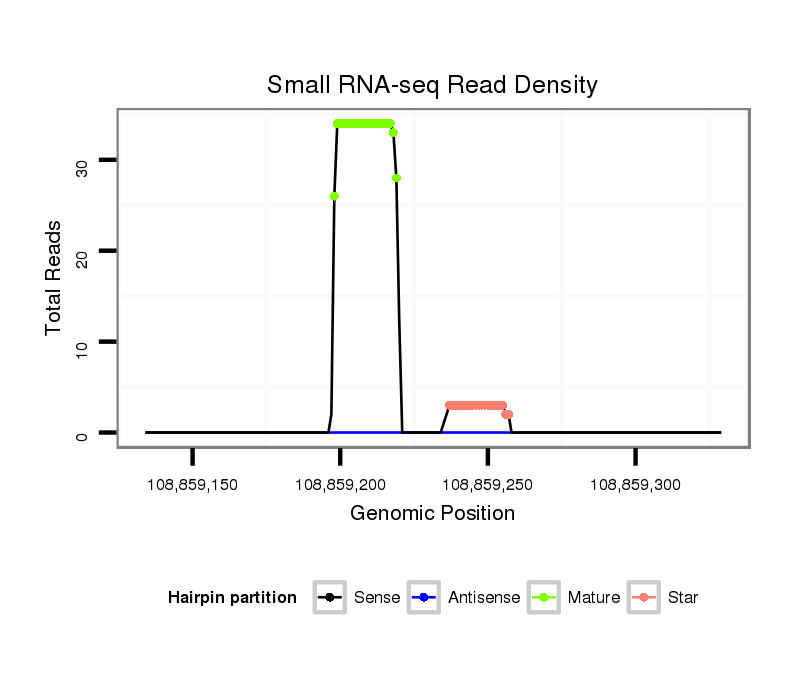
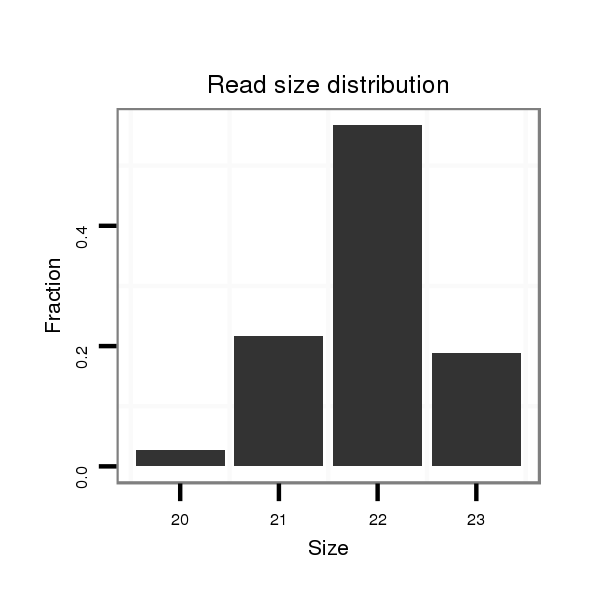
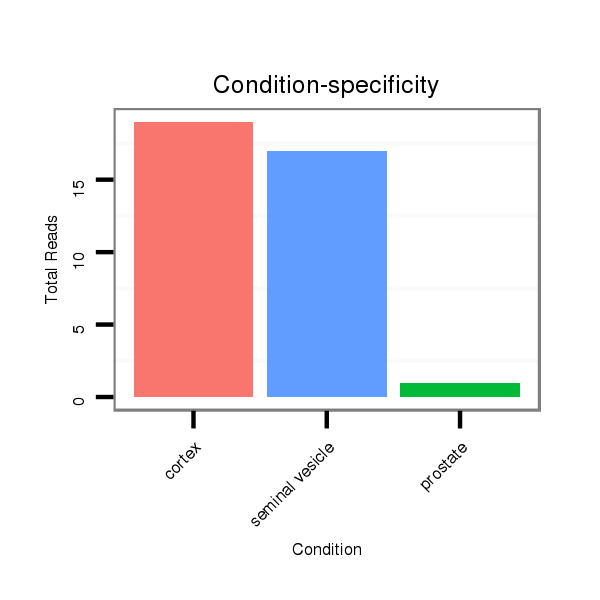
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116842 seminal vesicle |
SRR116841 prostate |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................GTAGATAAAATATTGGTACCTGT............................................................................................................. | 23 | 1 | 1 | 20.00 | 20 | 12 | 7 | 1 | 0 |
| ................................................................GTAGATAAAATATTGGTACCTG.............................................................................................................. | 22 | 0 | 1 | 13.00 | 13 | 6 | 7 | 0 | 0 |
| ................................................................GTAGATAAAATATTGGTACCTGA............................................................................................................. | 23 | 0 | 1 | 7.00 | 7 | 3 | 4 | 0 | 0 |
| .................................................................TAGATAAAATATTGGTACCTGA............................................................................................................. | 22 | 0 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 |
| .................................................................TAGATAAAATATTGGTACCTG.............................................................................................................. | 21 | 0 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 |
| ................................................................GTAGATAAAATATTGGTACCT............................................................................................................... | 21 | 0 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 |
| ...............................................................AGTAGATAAAATATTGGTACCT............................................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 |
| ................................................................GTAGATAAAATATTGGTACCTT.............................................................................................................. | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................AGTAGATAAAATATTGGTACCTGT............................................................................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................TTTCAATACTTTATCTGCTCT........................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................GTAGATAAAATCTTGGTACCTGT............................................................................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................GGTTTCAATACTTTATCTGCT.......................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................GTAGATAAAATTTTGGTACC................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................................................GTAGATAAAATATTTGTACCTGA............................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................TAGGTAAAATATTGGTACCTG.............................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GTTTCAATACTTTATCTGCTCT........................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................GTAGATAAAATATTGGTACC................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................................................GTAGATAAAATATTGGTACCTGTAA........................................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................GTAGATAAAATATTGTTACCTGT............................................................................................................. | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ................................................................GTAGATAAAATATTGGTACCTTT............................................................................................................. | 23 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................GGTTTCAATACTTTATCTGT........................................................................... | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ................................................................GTAGATAAAATATTGGTACCTATC............................................................................................................ | 24 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................GATAAAATATTGGTACCTGTA............................................................................................................ | 21 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................CGCTCTGAAGACAGAA.... | 16 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
TACACGACCCCCCGCCTCCTTCAAAGTCCATGGGTTACCGTCCGACGTAAACCCCCTTACTTCTCATCTATTTTATAACCATGGACTACTCACACTCCGGTCCAAAGTTATGAAATAGACGAGAAGTAAAAGGGTATAGATGAATGAAATCCATACCGGAGTAATTTTTTATATTTGAGAGACTTCTGTCTTGTAA
*********************************************************.(((((((((((((((((((((((.((((((.(((((.............))))).)))))).)))))))))))))))))))))))....************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:24 PM