
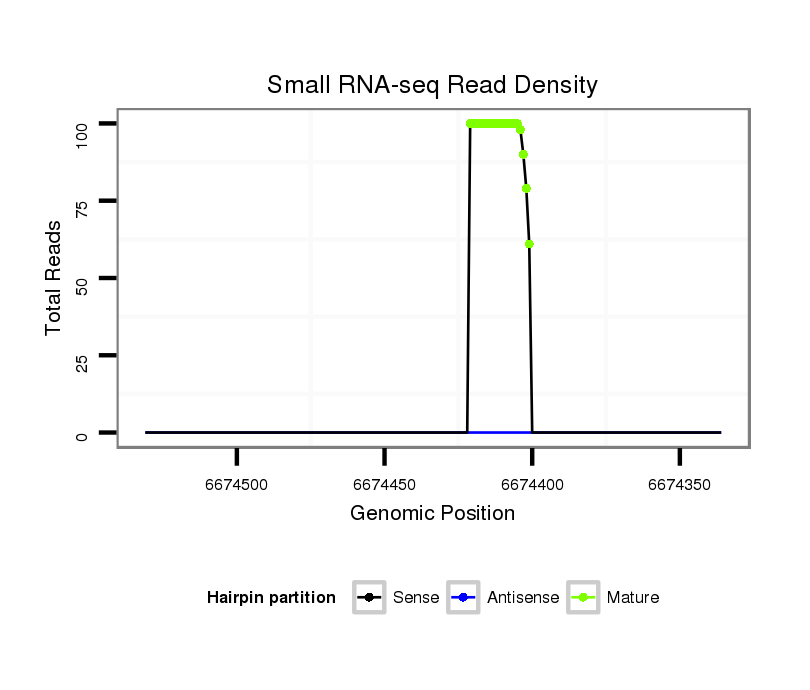
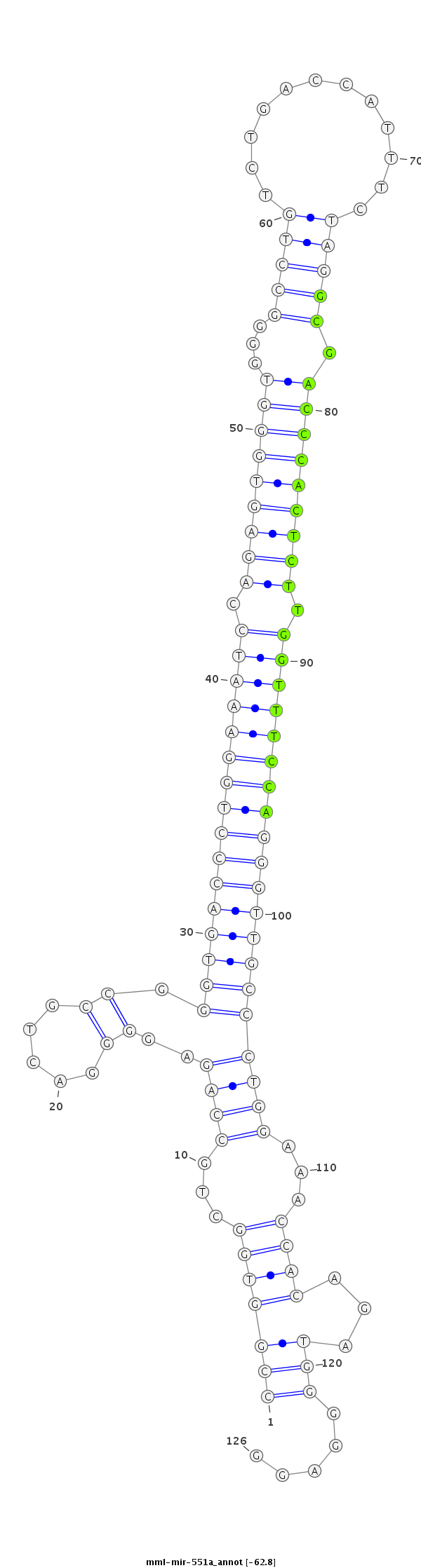
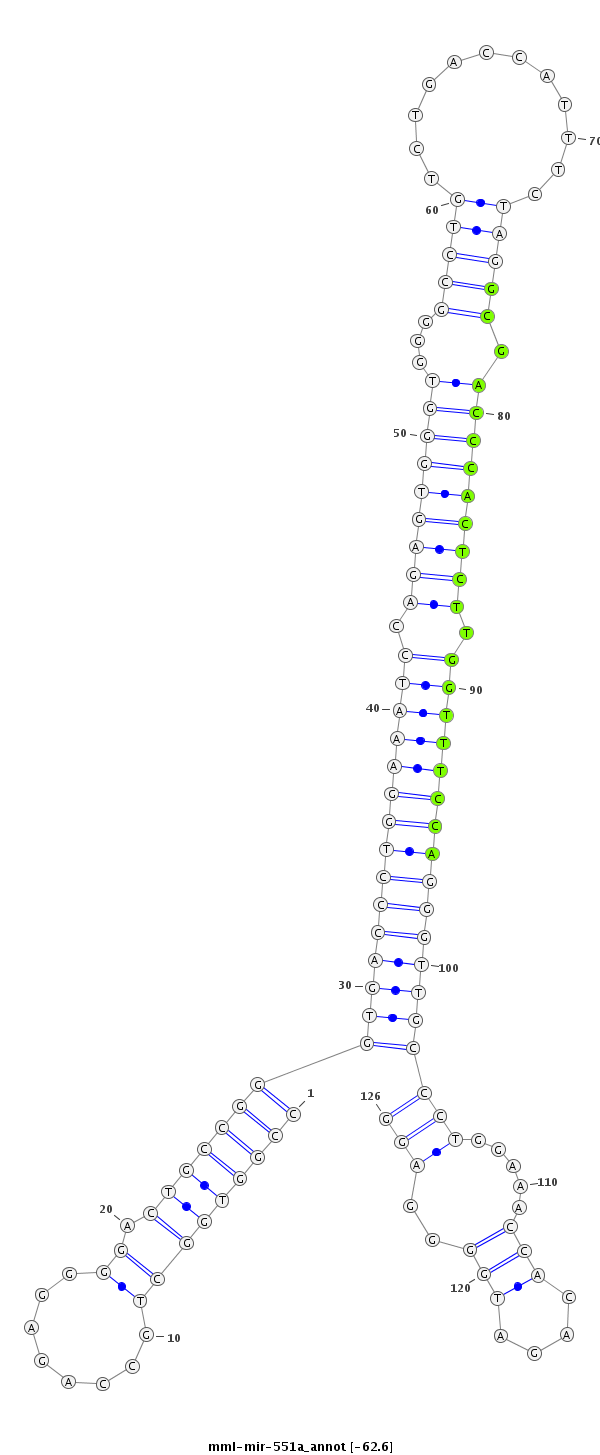
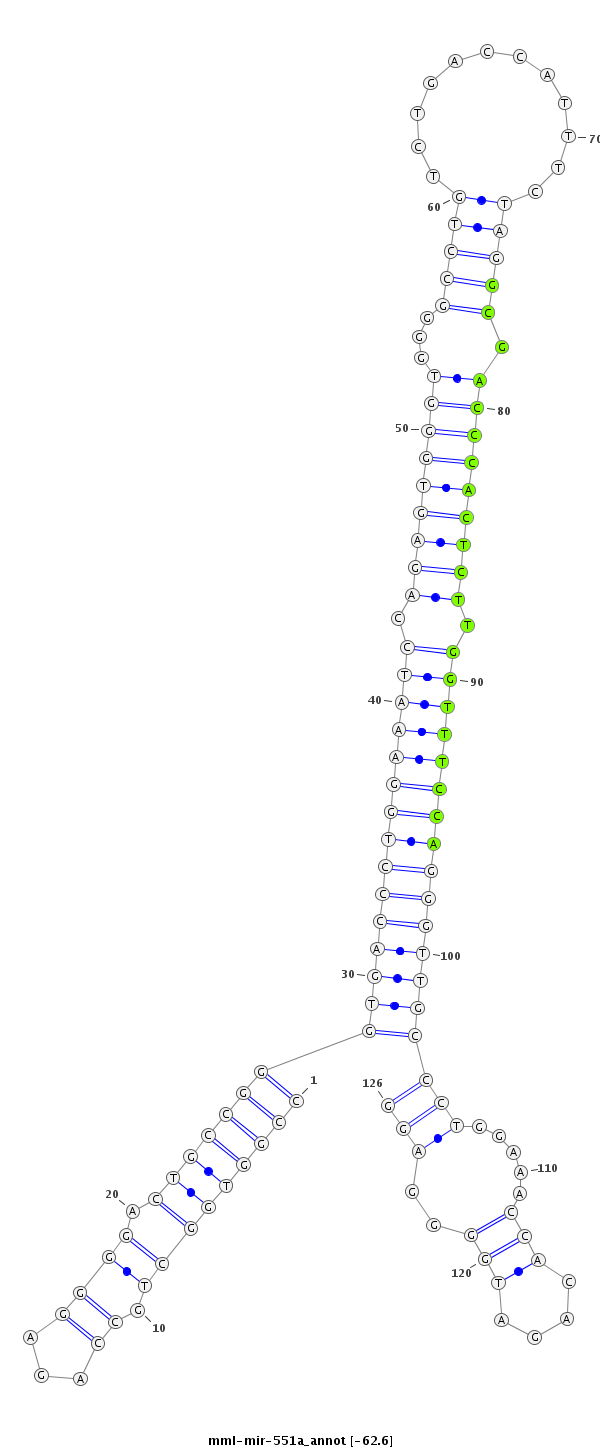
ID:mml-mir-551a |
Coordinate:chr1:6674386-6674481 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -62.8 | -62.6 | -62.6 |
 |
 |
 |
|
CTCGCGGCCAACCCCTGAGGCTGTTCCTGGCTGCTCCGGTGGCTGCCAGAGGGGACTGCCGGGTGACCCTGGAAATCCAGAGTGGGTGGGGCCTGTCTGACCATTTCTAGGCGACCCACTCTTGGTTTCCAGGGTTGCCCTGGAAACCACAGATGGGGAGGGGTTGATGGCACCCAGCCTCCCCCAAGCCTGGGAA
***********************************(((((((...((((.((......)).((((((((((((((((.(((((((((...(((((............))))).))))))))).))))))))))))))))))))...))))...))).....*********************************** |
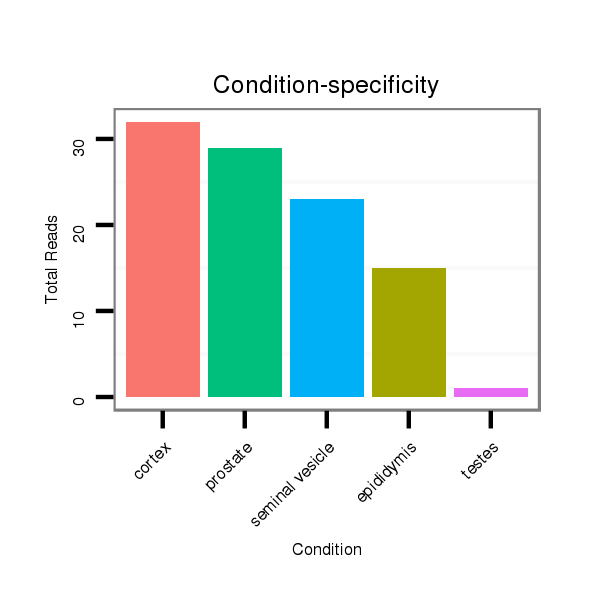
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116841 prostate |
SRR116842 seminal vesicle |
SRR116840 epididymis |
SRR116839 testes |
SRR116845 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|---|
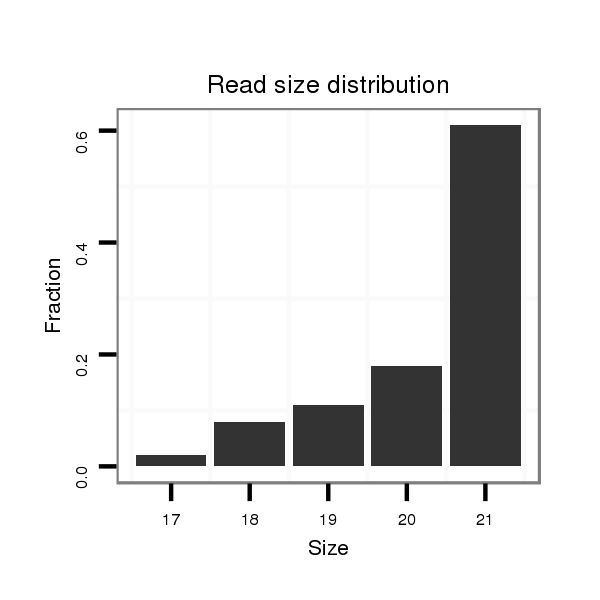
| ..............................................................................................................GCGACCCACTCTTGGTTTCCA................................................................. | 21 | 0 | 1 | 61.00 | 61 | 20 | 20 | 14 | 7 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCC.................................................................. | 20 | 0 | 1 | 18.00 | 18 | 4 | 5 | 3 | 5 | 1 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTC................................................................... | 19 | 0 | 1 | 11.00 | 11 | 5 | 4 | 1 | 1 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTT.................................................................... | 18 | 0 | 1 | 8.00 | 8 | 2 | 0 | 5 | 1 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCCAT................................................................ | 22 | 1 | 1 | 4.00 | 4 | 1 | 2 | 0 | 1 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGTTTTCCA................................................................. | 21 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTT..................................................................... | 17 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTA.................................................................... | 18 | 1 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTTC.................................................................. | 20 | 1 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCCC................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCT.................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCCATC............................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................GCGACCCACTCTTTGTTTCCA................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCCAA................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCGA................................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCCT................................................................. | 21 | 1 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCCCTCTTGGTTTCC.................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCA.................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTTGTTTCC.................................................................. | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTAA.................................................................. | 20 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTTCAT................................................................. | 21 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................GCGACCCACTCTTGGTTCCA.................................................................. | 20 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
GAGCGCCGGTTGGGGACTCCGACAAGGACCGACGAGGCCACCGACGGTCTCCCCTGACGGCCCACTGGGACCTTTAGGTCTCACCCACCCCGGACAGACTGGTAAAGATCCGCTGGGTGAGAACCAAAGGTCCCAACGGGACCTTTGGTGTCTACCCCTCCCCAACTACCGTGGGTCGGAGGGGGTTCGGACCCTT
***********************************(((((((...((((.((......)).((((((((((((((((.(((((((((...(((((............))))).))))))))).))))))))))))))))))))...))))...))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:02 PM