ID:mml-mir-548i |
Coordinate:chr3:9672271-9672328 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
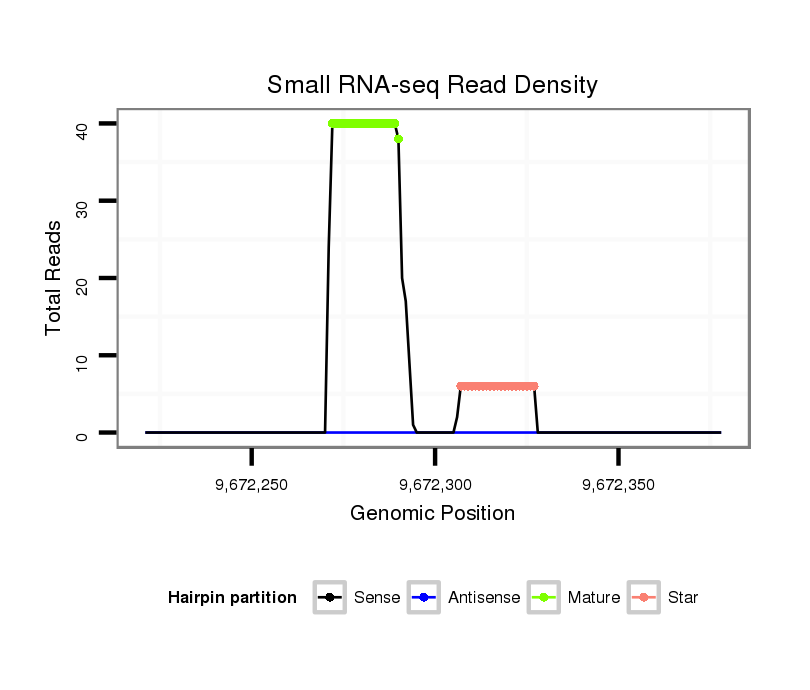
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
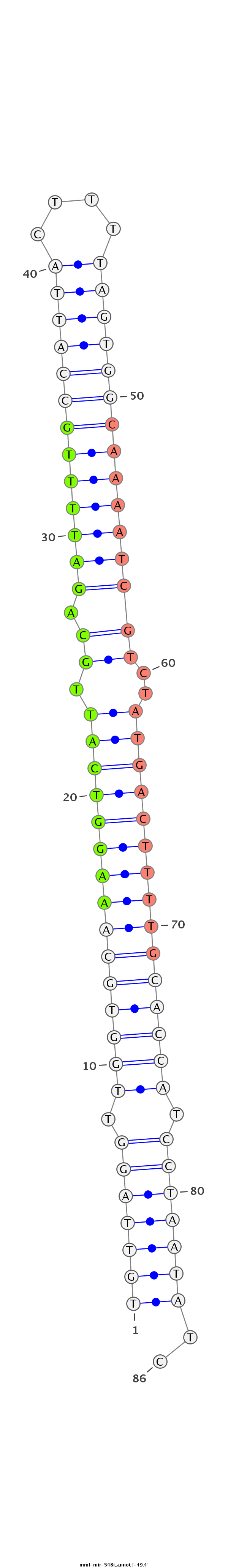
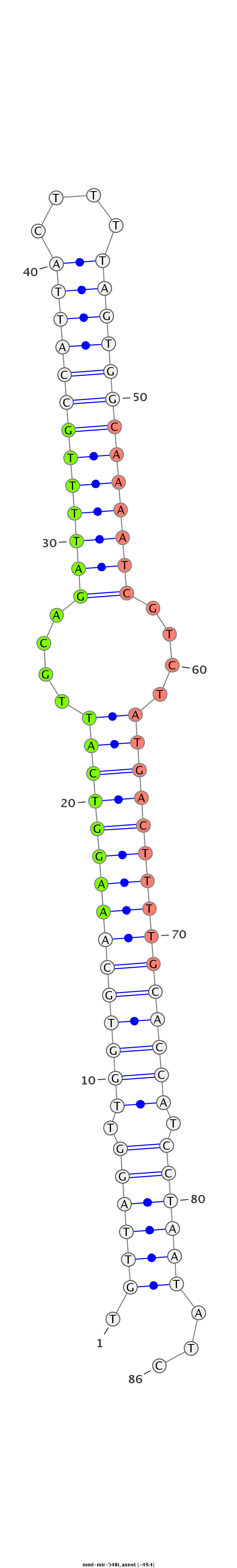
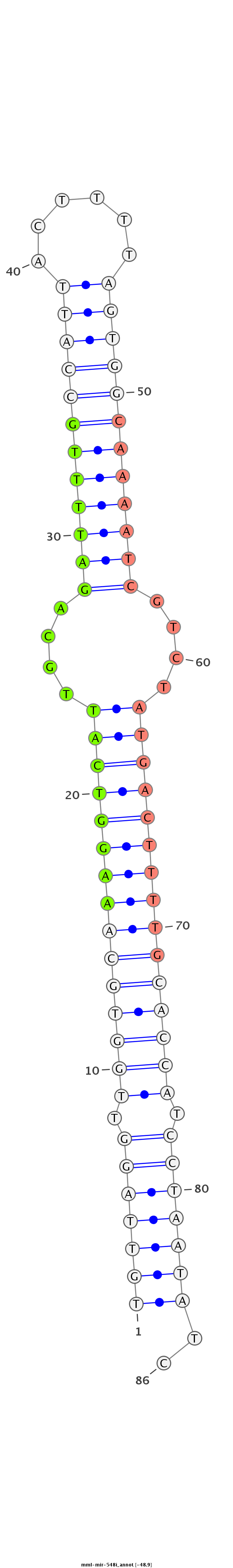
| -49.4 | -49.4 | -48.9 |
 |
 |
 |
|
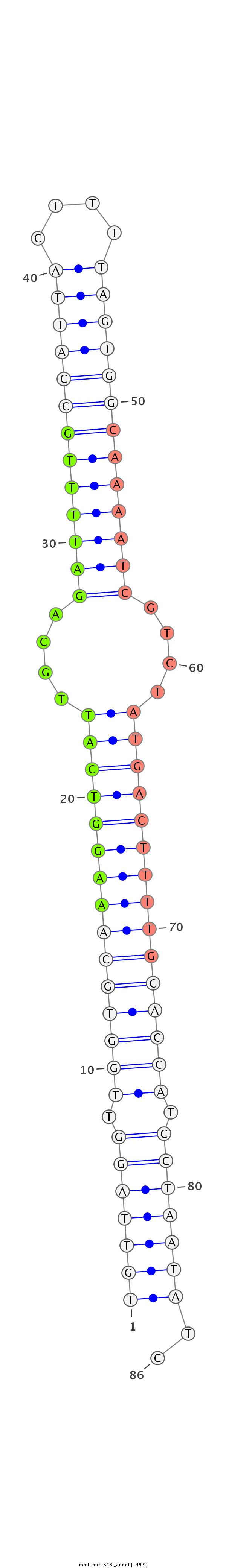
GGCAGCTGGGATCAGAAATACAAACGAACAAGTAACTGTTAGGTTGGTGCAAAGGTCATTGCAGATTTTGCCATTACTTTTAGTGGCAAAATCGTCTATGACTTTTGCACCATCCTAATATCTAGCTAACAGGTCTCTCAGCTGCTCATCTCTGTTCC
************************************(((((((.(((((((((((((((....(((((((((((((....)))))))))))))....))))))))))))))).)))))))..************************************ |
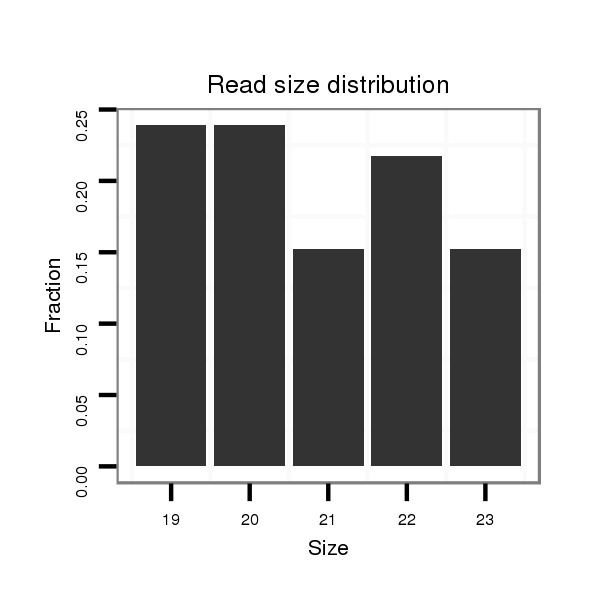
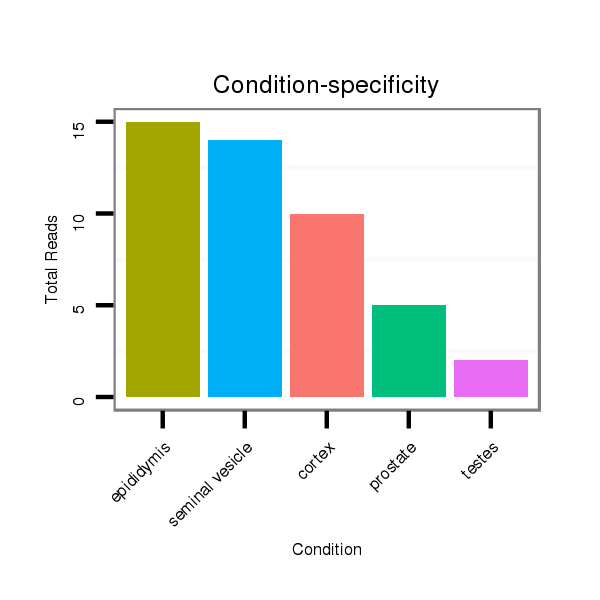
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116842 seminal vesicle |
SRR116843 cortex |
SRR116841 prostate |
SRR116845 epididymis |
SRR116839 testes |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................AAGGTCATTGCAGATTTTG........................................................................................ | 19 | 0 | 1 | 9.00 | 9 | 4 | 5 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTG........................................................................................ | 20 | 0 | 1 | 9.00 | 9 | 4 | 1 | 3 | 1 | 0 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTGCC...................................................................................... | 22 | 0 | 1 | 6.00 | 6 | 2 | 0 | 1 | 3 | 0 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTGCCA..................................................................................... | 23 | 0 | 1 | 6.00 | 6 | 1 | 3 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................CAAAATCGTCTATGACTTTTG................................................... | 21 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAATCGTCTATGACTTTTGATC................................................ | 24 | 2 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| ...................................................AAGGTCATTGCAGATTTTGC....................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| ...................................................AAGGTCATTGCAGATTTTGCCA..................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GCAAAATCGTCTATGACTTTTGATC................................................ | 25 | 2 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTT......................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................AAGGTCATTGCAGATTTTGCC...................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GCAAAATCGTCTATGACTTTTG................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................................................AAGGGCATTGCAGATTTTGCC...................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTGC....................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTGCCT..................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................AAGGTCATTGCAGATTTTGCCAT.................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAATCGTCTATGACTTTTGA.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................AAGGTCATTGCAGATTTTTCC...................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................AAAGGACATTGCAGATTTTGCCA..................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................CAGAATCGTCTATGACTTTTG................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................AAGGTCATTGCAGATTTTGA....................................................................................... | 20 | 1 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTT........................................................................................ | 20 | 1 | 3 | 0.67 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTGA....................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................AAGGTCATTGCAGATTTTGT....................................................................................... | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AAAGGTCGTTGCAGATTTTGA....................................................................................... | 21 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................AAGGTCATTGCAGATTTTTCA...................................................................................... | 21 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................AAGGTCATTGCAGATTTTGAAA..................................................................................... | 22 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AAAGGTCATTGCAGATTTTGATC..................................................................................... | 23 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
CCGTCGACCCTAGTCTTTATGTTTGCTTGTTCATTGACAATCCAACCACGTTTCCAGTAACGTCTAAAACGGTAATGAAAATCACCGTTTTAGCAGATACTGAAAACGTGGTAGGATTATAGATCGATTGTCCAGAGAGTCGACGAGTAGAGACAAGG
************************************(((((((.(((((((((((((((....(((((((((((((....)))))))))))))....))))))))))))))).)))))))..************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116844 epididymis |
|---|---|---|---|---|---|---|
| ................CTATGGTTGCTTGTTCATT........................................................................................................................... | 19 | 2 | 15 | 0.07 | 1 | 1 |
Generated: 09/01/2015 at 12:20 PM