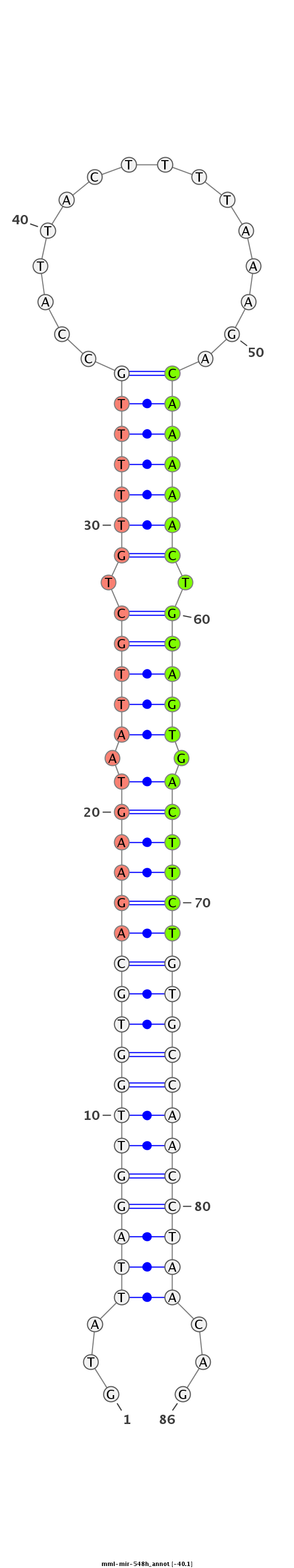
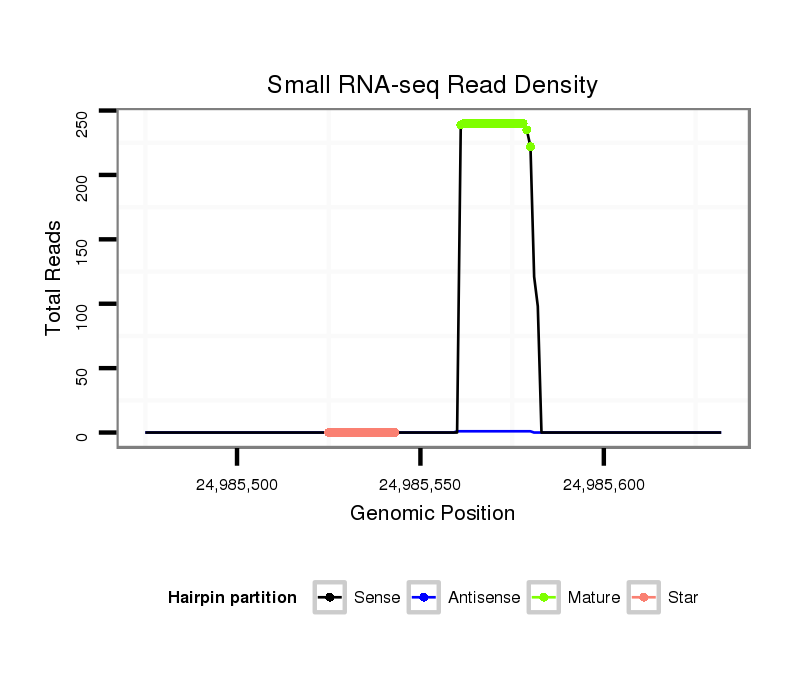
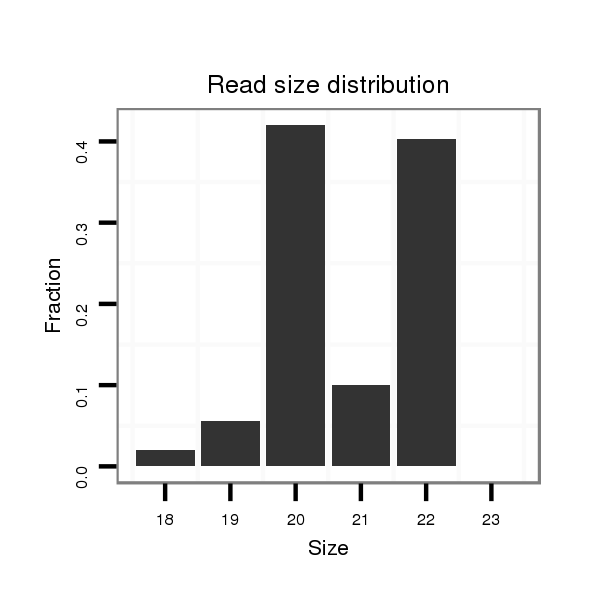
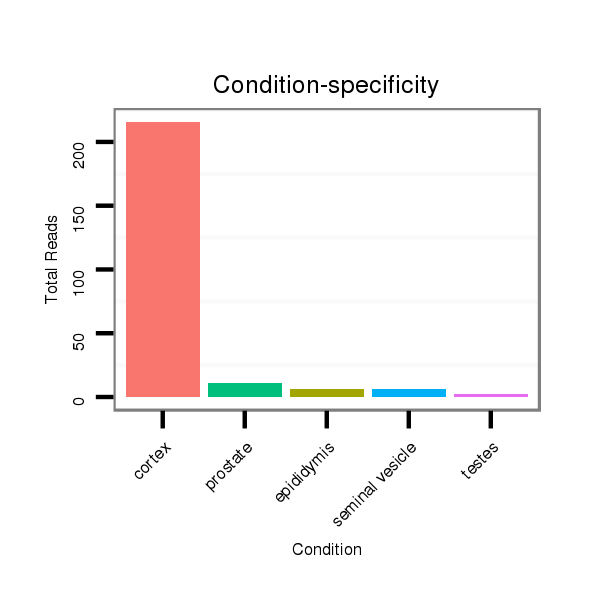
ID:mml-mir-548h |
Coordinate:chr13:24985525-24985582 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.1 | -39.2 | -39.1 |
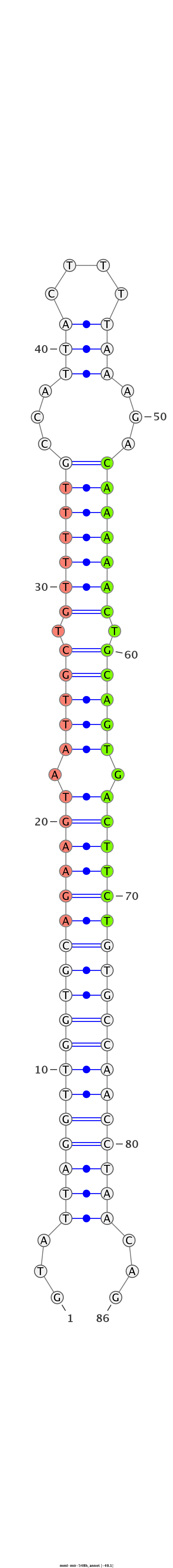 |
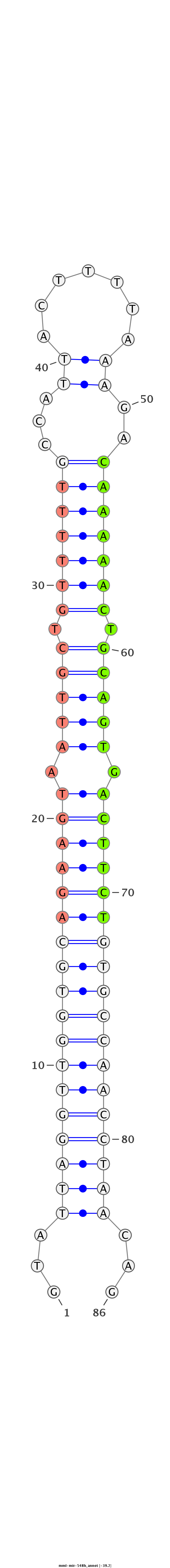 |
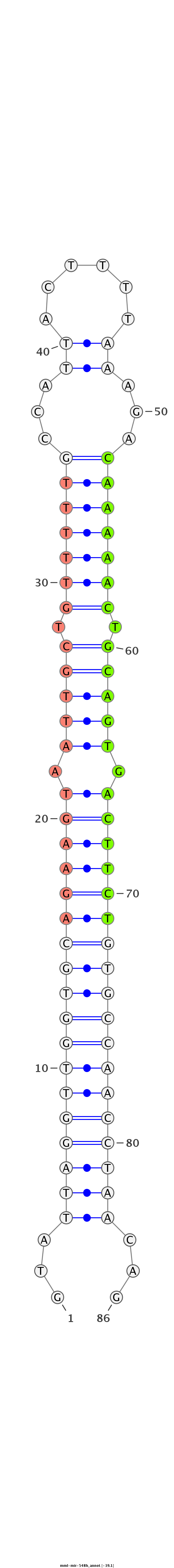 |
| mature | star |
|
GTCACTTTCTGTTTATCTGTGCAATGAGGATGGTTGTATTAGGTTGGTGCAGAAGTAATTGCTGTTTTTGCCATTACTTTTAAAGACAAAAACTGCAGTGACTTCTGTGCCAACCTAACAGTATCTACGTAAGGGGTTGTTCTGAGGTTTACATGAGT
***********************************...((((((((((((((((((.(((((.(((((((................))))))).))))).))))))))))))))))))...************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116841 prostate |
SRR116840 epididymis |
SRR116842 seminal vesicle |
SRR116839 testes |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................CAAAAACTGCAGTGACTTCT.................................................... | 20 | 0 | 1 | 101.00 | 101 | 91 | 3 | 3 | 3 | 1 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTCTGT.................................................. | 22 | 0 | 1 | 97.00 | 97 | 86 | 6 | 2 | 2 | 1 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTCTG................................................... | 21 | 0 | 1 | 23.00 | 23 | 21 | 0 | 1 | 1 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTC..................................................... | 19 | 0 | 1 | 13.00 | 13 | 12 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTT...................................................... | 18 | 0 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTCTTT.................................................. | 22 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAGCTGCAGTGACTTCTGT.................................................. | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGCGACTTCT.................................................... | 20 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTCTT................................................... | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAGACTGCAGTGACTTCT.................................................... | 20 | 1 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGCGACTTCTGT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCATTGACTTCTG................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCATTGACTTCTGT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTCTGTATC............................................... | 25 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................AAAAACTGCAGTGACTTCTGT.................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAGAAACTGCAGTGACTTCTGTA................................................. | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGGCTTCT.................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGAAGTAATTGCTGTTTTT......................................................................................... | 19 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGAAGTAATTGCTGTTTTTGCCA..................................................................................... | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTCTGAA................................................. | 23 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGAAGTAATTGCTGTTTTTG........................................................................................ | 20 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGAAGTAATTTCTGTTTTTGC....................................................................................... | 21 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTTTGT.................................................. | 22 | 1 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................AGAAGTAATTGCTGTTTTTCCT...................................................................................... | 22 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CAAAAACTGCAGTGACTTTTG................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CAGTGAAAGACAAATAGACACGTTACTCCTACCAACATAATCCAACCACGTCTTCATTAACGACAAAAACGGTAATGAAAATTTCTGTTTTTGACGTCACTGAAGACACGGTTGGATTGTCATAGATGCATTCCCCAACAAGACTCCAAATGTACTCA
*************************************...((((((((((((((((((.(((((.(((((((................))))))).))))).))))))))))))))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
|---|---|---|---|---|---|---|
| .....................................................................................TGTTTTTGACGTCACTGAAGA.................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 |
| ...................................................................................ACTGTTTTTGACGTCACTGAAGA.................................................... | 23 | 1 | 3 | 0.33 | 1 | 1 |
Generated: 09/01/2015 at 12:09 PM