ID:mml-mir-548g |
Coordinate:chr13:17105868-17105922 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -45.3 |
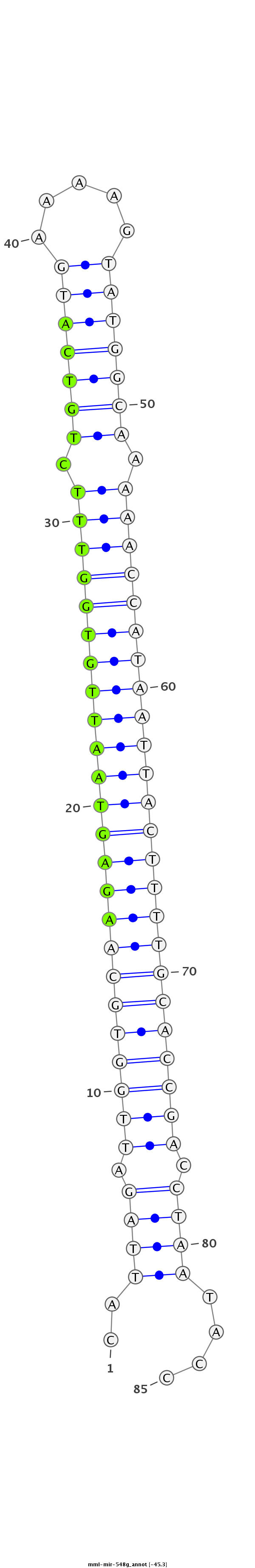 |
|
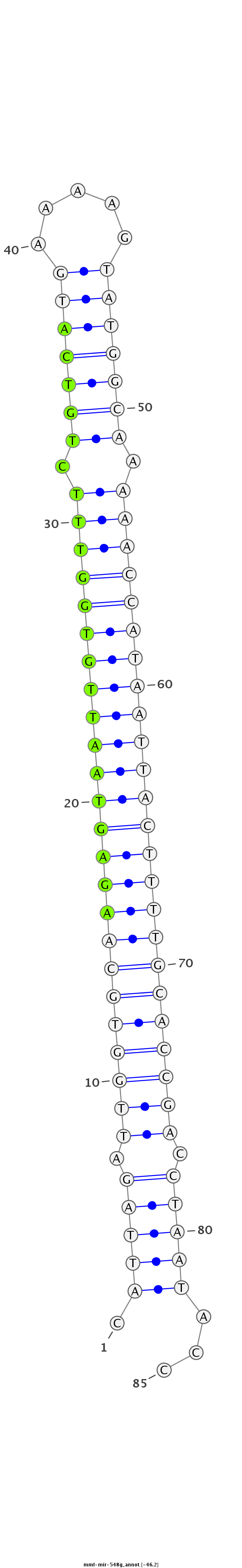
CAACCTCAATCTTACAACTCCAAGGAACTACATGCCATTAGATTGGTGCAAGAGTAATTGTGGTTTCTGTCATGAAAAGTATGGCAAAAACCATAATTACTTTTGCACCGACCTAATACCAACAAGCAGATAAGCTTGGAAAACACCCCTGAACT
***********************************.(((((.((((((((((((((((((((((((.(((((((.....))))))).)))))))))))))))))))))))).)))))...*********************************** |
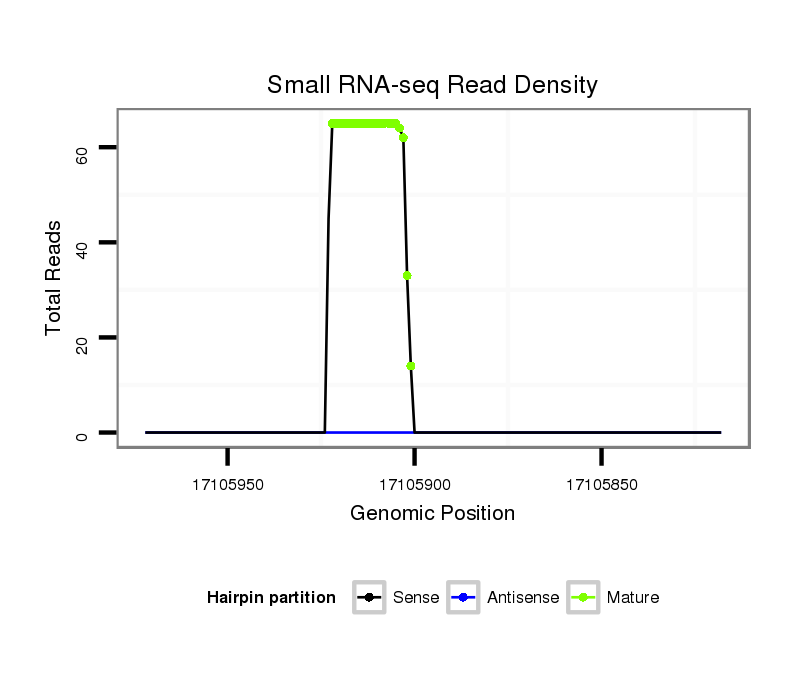
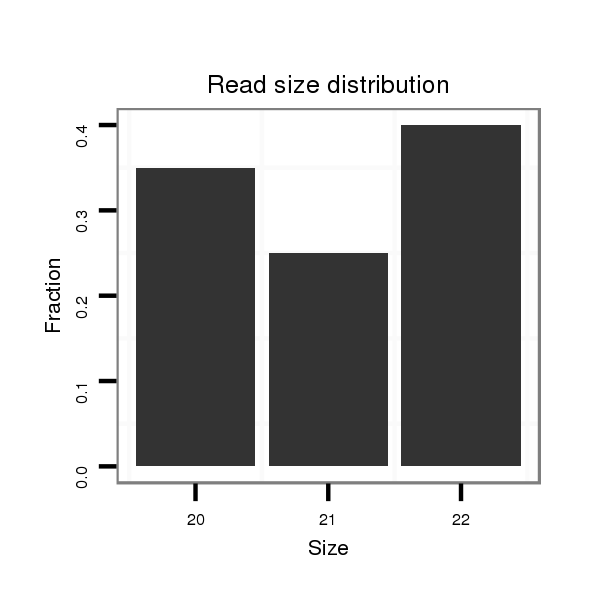
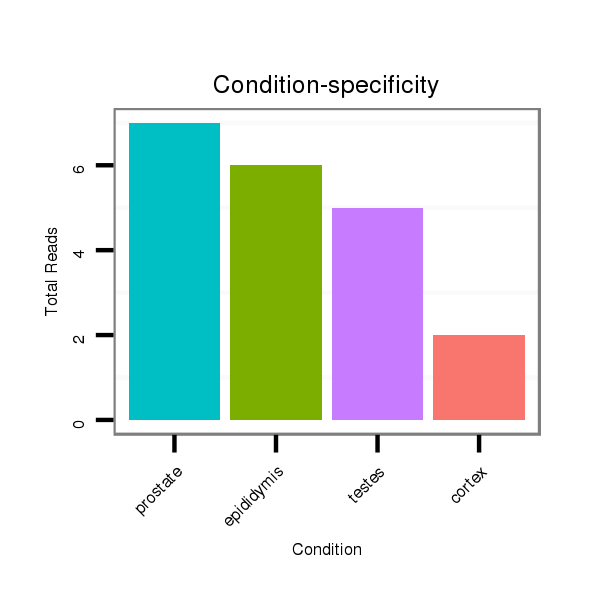
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116839 testes |
SRR116841 prostate |
SRR116840 epididymis |
SRR116842 seminal vesicle |
SRR116843 cortex |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................AAGAGTAATTGTGGTTTCTGT..................................................................................... | 21 | 0 | 1 | 22.00 | 22 | 7 | 8 | 5 | 1 | 1 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTGTC.................................................................................... | 22 | 0 | 1 | 14.00 | 14 | 7 | 3 | 2 | 2 | 0 | 0 |
| ..................................................AGAGTAATTGTGGTTTCTGTCA................................................................................... | 22 | 0 | 1 | 8.00 | 8 | 1 | 4 | 3 | 0 | 0 | 0 |
| ..................................................AGAGTAATTGTGGTTTCTGT..................................................................................... | 20 | 0 | 1 | 7.00 | 7 | 2 | 2 | 1 | 0 | 2 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTGTCA................................................................................... | 23 | 0 | 1 | 6.00 | 6 | 4 | 0 | 2 | 0 | 0 | 0 |
| ..................................................AGAGTAATTGTGGTTTCTGTC.................................................................................... | 21 | 0 | 1 | 5.00 | 5 | 2 | 1 | 2 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTGTA.................................................................................... | 22 | 1 | 1 | 4.00 | 4 | 3 | 0 | 0 | 1 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTGTCT................................................................................... | 23 | 1 | 2 | 2.50 | 5 | 2 | 3 | 0 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTTTC.................................................................................... | 22 | 1 | 2 | 2.50 | 5 | 3 | 0 | 2 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTGTAA................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTG...................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTTT..................................................................................... | 21 | 1 | 4 | 1.50 | 6 | 2 | 1 | 2 | 0 | 1 | 0 |
| .................................................AAGAGTAATTGTGGTTTCT....................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTTTCA................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGAGTAATTGTGGTTTCTTTCA................................................................................... | 22 | 1 | 2 | 1.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 |
| ..................................................AGAGTAATTGTGGTTTCTTTC.................................................................................... | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTTTCT................................................................................... | 23 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTT...................................................................................... | 20 | 1 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTTTA.................................................................................... | 22 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................AGAGTAATTGTGGTTTCTTT..................................................................................... | 20 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................AAGAGTAATTGTGGTTTCTGTATC.................................................................................. | 24 | 3 | 13 | 0.15 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| .................................................AGGAGTAATTGTGGTTTCTTT..................................................................................... | 21 | 2 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................AGAGTAATTGTGGTTTCTGAA.................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GTTGGAGTTAGAATGTTGAGGTTCCTTGATGTACGGTAATCTAACCACGTTCTCATTAACACCAAAGACAGTACTTTTCATACCGTTTTTGGTATTAATGAAAACGTGGCTGGATTATGGTTGTTCGTCTATTCGAACCTTTTGTGGGGACTTGA
***********************************.(((((.((((((((((((((((((((((((.(((((((.....))))))).)))))))))))))))))))))))).)))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:08 PM