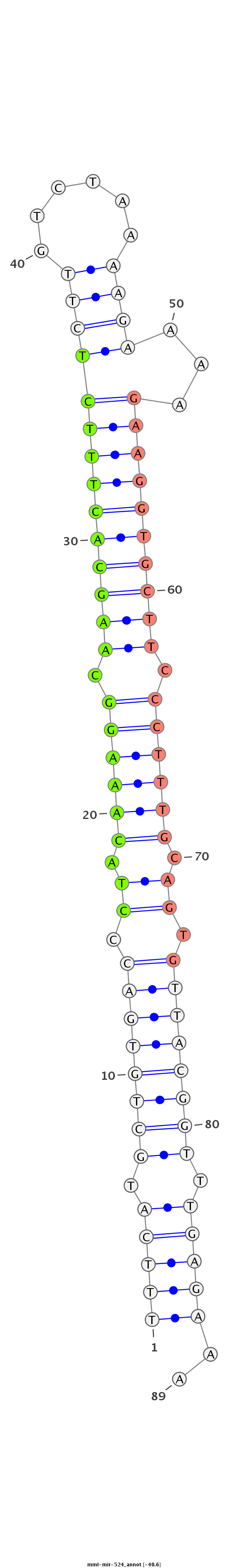


ID:mml-mir-524 |
Coordinate:chr19:59594913-59594997 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.3 | -39.7 |
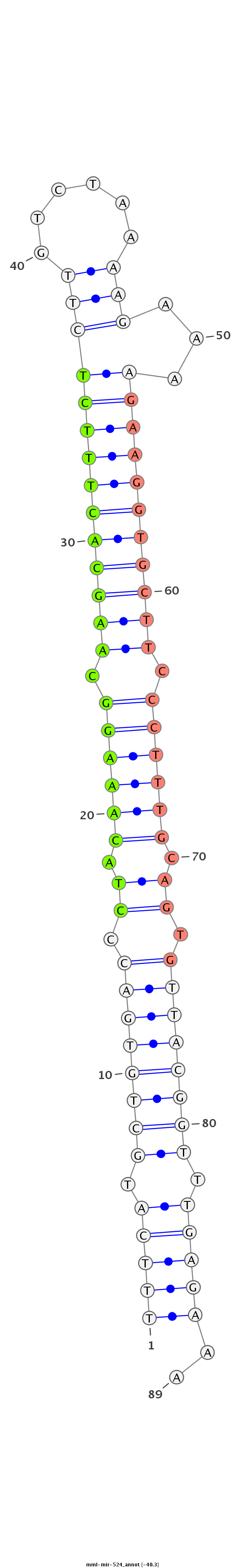 |
 |
| mature | star |
|
GGTTAAGGAAAATTTCCACAAGAAATGCAGAGTACTGGAGCAAGAAGATTTCATGCTGTGACCCTACAAAGGCAAGCACTTTCTCTTGTCTAAAAGAAAAGAAGGTGCTTCCCTTTGCAGTGTTACGGTTTGAGAAAAGCAACGTTGAAGTCAATGCTGATCTTGGTAATACATTTGCAGCTCAT
************************************************(((((.((((((((.((.((((((.((((((((((((((......))))...)))))))))).)))))).)).)))))))).)))))..************************************************ |
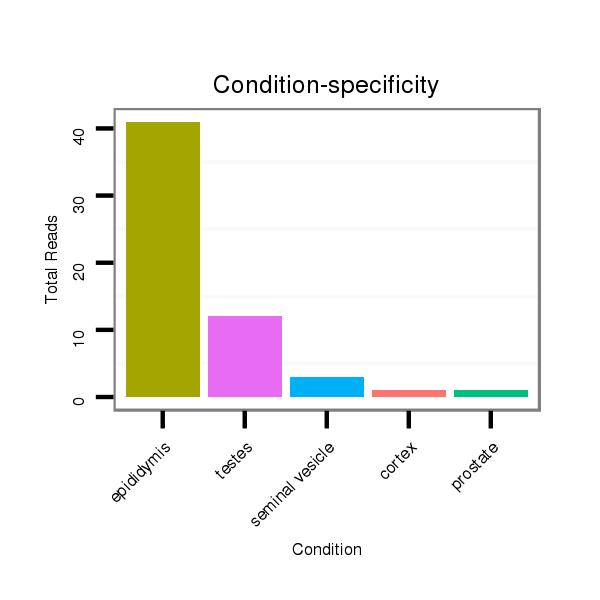
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116839 testes |
SRR116842 seminal vesicle |
SRR116841 prostate |
SRR116843 cortex |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................CTACAAAGGCAAGCACTTTCT..................................................................................................... | 21 | 0 | 2 | 41.50 | 83 | 66 | 13 | 4 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTTCTC.................................................................................................... | 22 | 0 | 2 | 4.50 | 9 | 1 | 4 | 2 | 0 | 2 |
| ...............................................................CTACAAAGGCAAGCACTTTCTCT................................................................................................... | 23 | 0 | 2 | 3.00 | 6 | 3 | 3 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTTC...................................................................................................... | 20 | 0 | 2 | 3.00 | 6 | 5 | 0 | 0 | 1 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTT....................................................................................................... | 19 | 0 | 2 | 2.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTTCA..................................................................................................... | 21 | 1 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...........................................GAAGATTTCATGCTGTGACC.......................................................................................................................... | 20 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTTCTCA................................................................................................... | 23 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................GAAGGTGCTTCCCTTTGCAGTG............................................................... | 22 | 0 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTTAA..................................................................................................... | 21 | 2 | 6 | 0.50 | 3 | 3 | 0 | 0 | 0 | 0 |
| ................................................................TACAAAGGCAAGCACTTTCTCT................................................................................................... | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTTCTCTT.................................................................................................. | 24 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................TACAAAGGCAAGCACTTTCT..................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................TACAAAGGCAAGCACTTTCTC.................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCGAGCACTTTCT..................................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTT........................................................................................................ | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................GAAGGTGCTTCCCTTTGCAGTGT.............................................................. | 23 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................CTAGAAAGGCAAGCACTTTCT..................................................................................................... | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................CTACAAAGGCAAGCACTTTCAT.................................................................................................... | 22 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................GAAGGTGCTTCCCTTTGCAGTTA.............................................................. | 23 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................CTACCAAGGCAAGCCCTTTCT..................................................................................................... | 21 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
|
CCAATTCCTTTTAAAGGTGTTCTTTACGTCTCATGACCTCGTTCTTCTAAAGTACGACACTGGGATGTTTCCGTTCGTGAAAGAGAACAGATTTTCTTTTCTTCCACGAAGGGAAACGTCACAATGCCAAACTCTTTTCGTTGCAACTTCAGTTACGACTAGAACCATTATGTAAACGTCGAGTA
************************************************(((((.((((((((.((.((((((.((((((((((((((......))))...)))))))))).)))))).)).)))))))).)))))..************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:18 PM