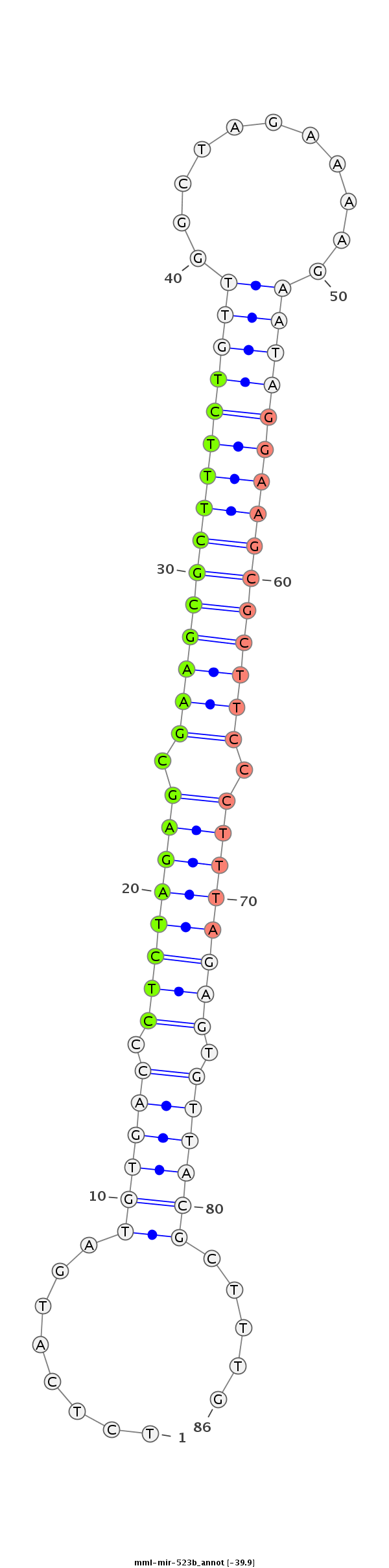
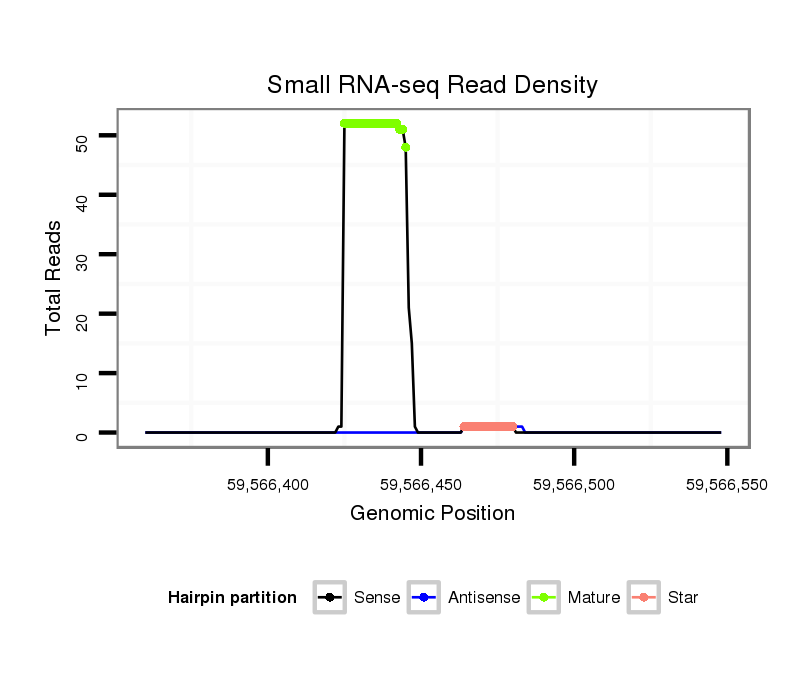
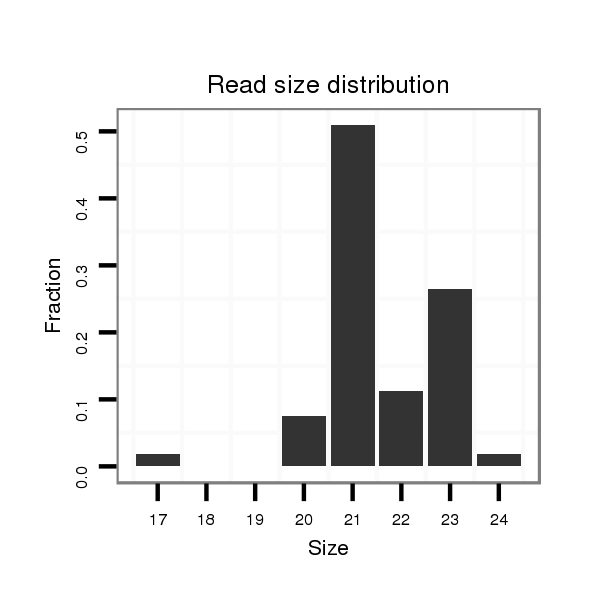
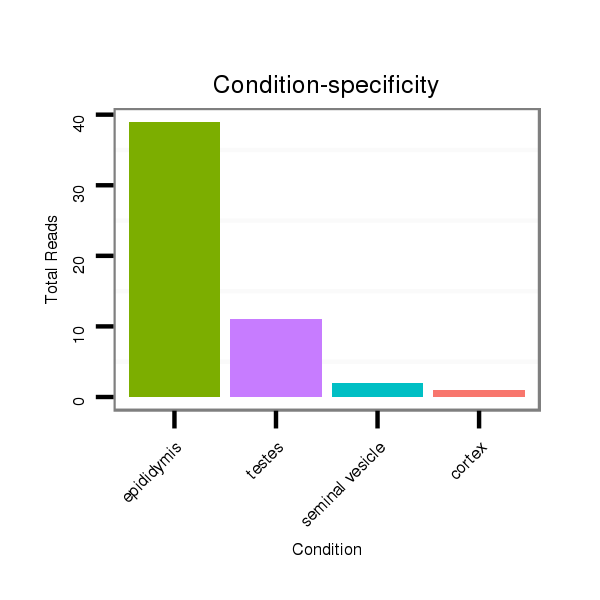
ID:mml-mir-523b |
Coordinate:chr19:59566410-59566498 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -39.3 | -38.9 |
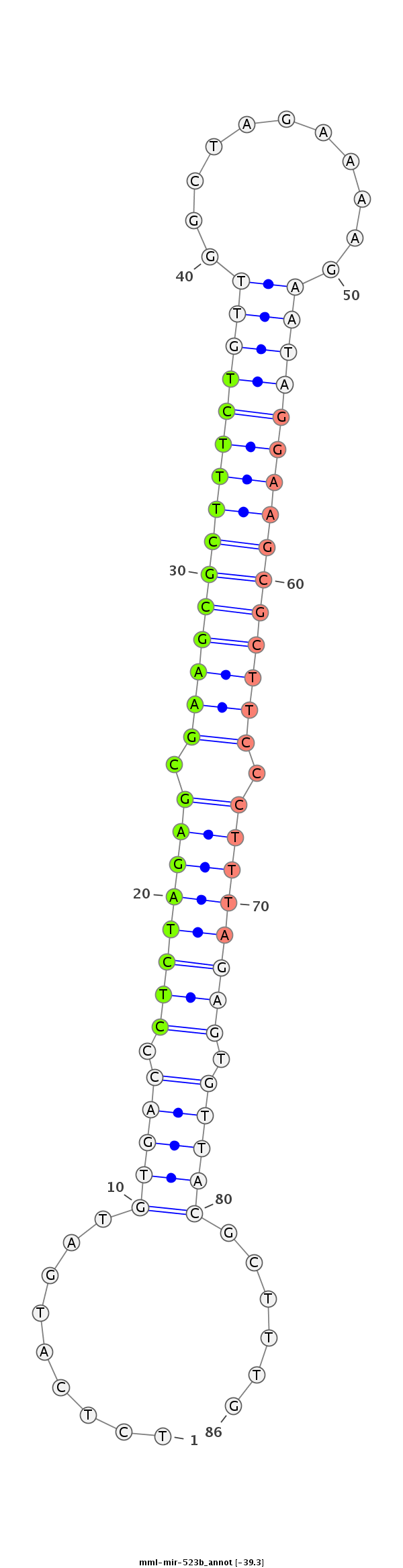 |
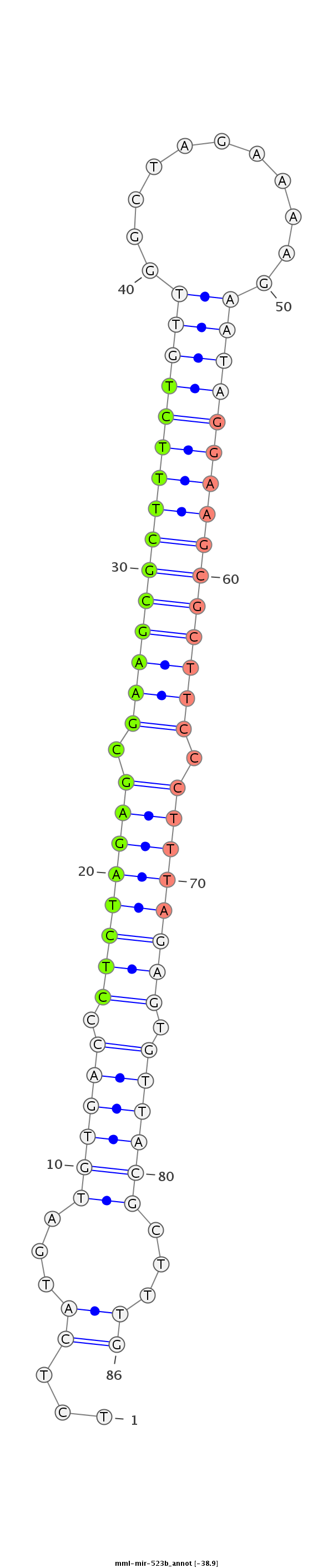 |
|
CCAGTGAAGGAAGATTCCAACAAAAAACCCAGAGTGCTGGAGCAAGAAGATCTCATGATGTGACCCTCTAGAGCGAAGCGCTTTCTGTTGGCTAGAAAAGAATAGGAAGCGCTTCCCTTTAGAGTGTTACGCTTTGAGAAAAGCAACGCGGATCTTGGTAATACACTTGCAGAGCGTGCTTATAATCAA
**************************************************........((((((.((((((((.(((((((((((((((...........))))))))))))))).)))))))).)))))).....***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116839 testes |
SRR116842 seminal vesicle |
SRR116841 prostate |
SRR116843 cortex |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................CTCTAGAGCGAAGCGCTTTCT....................................................................................................... | 21 | 0 | 1 | 27.00 | 27 | 24 | 1 | 2 | 0 | 0 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTCTGT..................................................................................................... | 23 | 0 | 1 | 14.00 | 14 | 7 | 6 | 0 | 0 | 1 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTCTG...................................................................................................... | 22 | 0 | 1 | 6.00 | 6 | 3 | 3 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGAGT................................................................ | 21 | 1 | 1 | 5.00 | 5 | 3 | 1 | 0 | 1 | 0 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTCTT...................................................................................................... | 22 | 1 | 1 | 3.00 | 3 | 0 | 2 | 0 | 1 | 0 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTC........................................................................................................ | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTCTTT..................................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CCCTCTAGAGCGAAGCGCTT.......................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCCCTTTAGAA................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CTCTAGAGCGTAGCGCTTTCTGT..................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTCTGTT.................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGAGTAA.............................................................. | 23 | 3 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGAGTGTA............................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGAGTG............................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTA........................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CTCTAGAGCGACGCGCTTTCT....................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCCCTTTAGAGTGA.............................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................CTCTAGAGCGAAGCGCTTTCTGTA.................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGAG................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TTCTAGAGCGAAGCGCTTTCTGT..................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................TTCTAGAGCGAAGCGCTTTCT....................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCCCTTTA.................................................................... | 17 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGATTG............................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGAGTA............................................................... | 22 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................GAAGCGCTTCTCTTTAGAGTGA.............................................................. | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................TGAAGCGCTTCTCTTTAGAGT................................................................ | 21 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................GGAAGCGCTTCTCTTTAGAGTAG.............................................................. | 23 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AAAACCAACGCGGATC................................... | 16 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................AAAAAACCAACGCGGATC................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GGTCACTTCCTTCTAAGGTTGTTTTTTGGGTCTCACGACCTCGTTCTTCTAGAGTACTACACTGGGAGATCTCGCTTCGCGAAAGACAACCGATCTTTTCTTATCCTTCGCGAAGGGAAATCTCACAATGCGAAACTCTTTTCGTTGCGCCTAGAACCATTATGTGAACGTCTCGCACGAATATTAGTT
*****************************************************........((((((.((((((((.(((((((((((((((...........))))))))))))))).)))))))).)))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116839 testes |
|---|---|---|---|---|---|---|---|
| ........................................................................................................CCTTCGCGAAGGGAAATCTC................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................................................................AGCTTTCGCGAAGGGAAATCTC................................................................. | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 |
Generated: 09/01/2015 at 12:17 PM