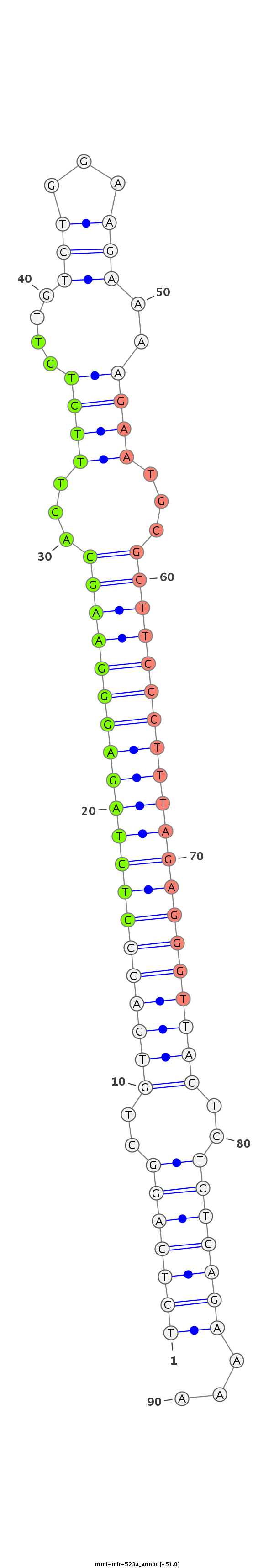
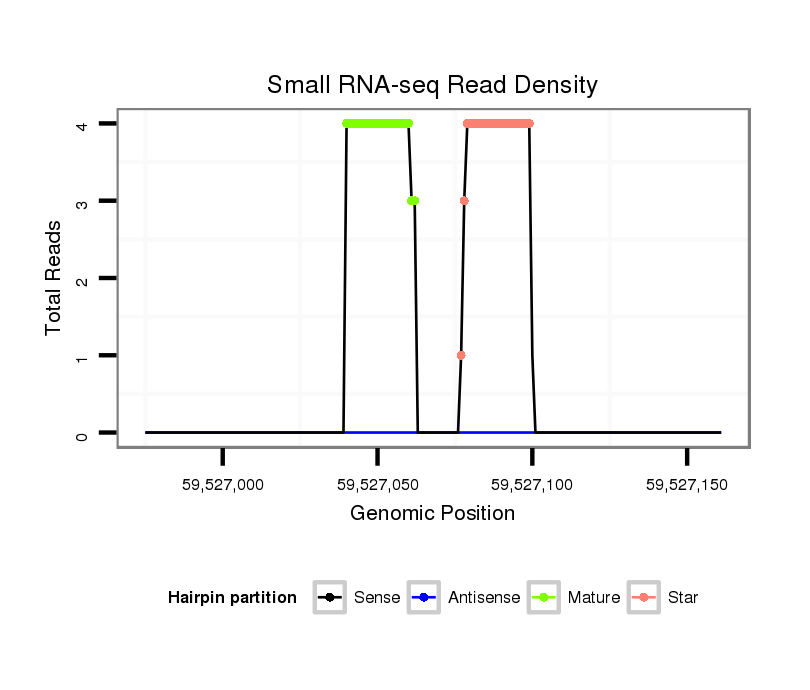

ID:mml-mir-523a |
Coordinate:chr19:59527025-59527111 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -50.1 |
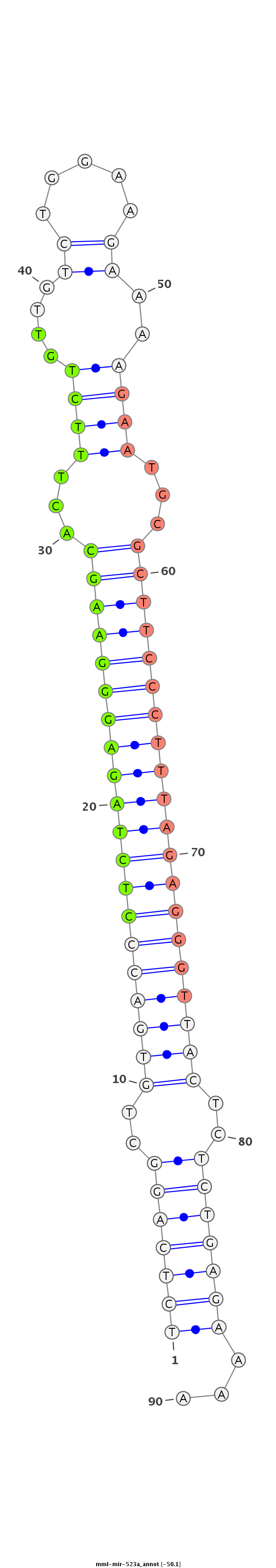 |
|
CTGGTCAAGGAAAATTTCTACAAAAAACCCAGGGTGCTGGAGCAAGAAGATCTCAGGCTGTGACCCTCTAGAGGGAAGCACTTTCTGTTGTCTGGAAGAAAAGAATGCGCTTCCCTTTAGAGGGTTACTCTCTGAGAAAAGCAATGTTGAAGTTGATGCAGATCTTGGTAATATATTTGCAGAGCAT
**************************************************(((((((..((((((((((((((((((((...((((....(((...)))..))))...))))))))))))))))))))..)))))))...*********************************************** |
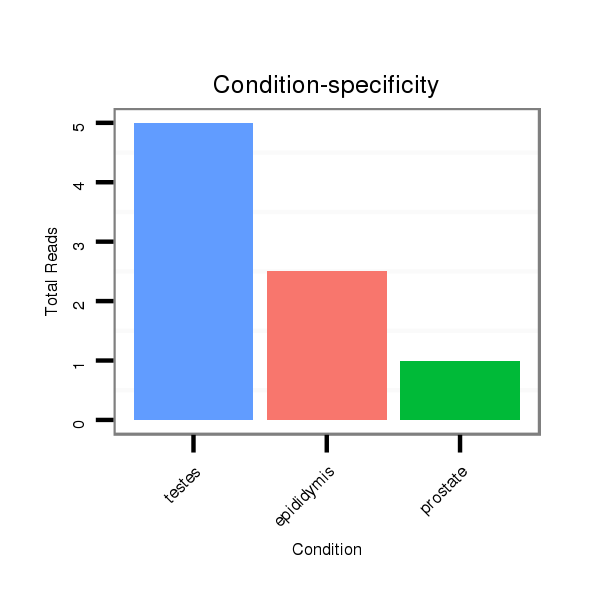
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116839 testes |
SRR116840 epididymis |
SRR116841 prostate |
SRR116842 seminal vesicle |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................CTCTAGAGGGAAGCACTTTCTGT................................................................................................... | 23 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 |
| .......................................................................................................AATGCGCTTCCCTTTAGAGGGC.............................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTCTTT................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................GAATGCGCTTCCCTTTAGAGGGT.............................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................AATGCGCTTCCCTTTAGAGGGTT............................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................AATGCGCTTCCCTTTAGAGGGT.............................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................ATGCGCTTCCCTTTAGAGGGT.............................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTCTGTAA................................................................................................. | 25 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................ATGCGCTTCCCTTTAGAGGGTTT............................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .......................................................................................................AATGCGCTTCCCTTTAGAGGGTAT............................................................ | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................AATGCGCTTCCCTTTAGAGGGTTT............................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTCT..................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTT...................................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................TCTAGAGGGAAGCACTTTCT..................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .................................................................TTCTAGAGGGAAGCACTTTCTGTT.................................................................................................. | 24 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ............................................AGAAGATCTCAGGCTGTGAC........................................................................................................................... | 20 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 |
|
GACCAGTTCCTTTTAAAGATGTTTTTTGGGTCCCACGACCTCGTTCTTCTAGAGTCCGACACTGGGAGATCTCCCTTCGTGAAAGACAACAGACCTTCTTTTCTTACGCGAAGGGAAATCTCCCAATGAGAGACTCTTTTCGTTACAACTTCAACTACGTCTAGAACCATTATATAAACGTCTCGTA
***********************************************(((((((..((((((((((((((((((((...((((....(((...)))..))))...))))))))))))))))))))..)))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
|---|---|---|---|---|---|---|
| ...................................................................................................TTTCTTTCGCGAAGGGAAATCTC................................................................. | 23 | 1 | 4 | 0.25 | 1 | 1 |
Generated: 09/01/2015 at 12:16 PM