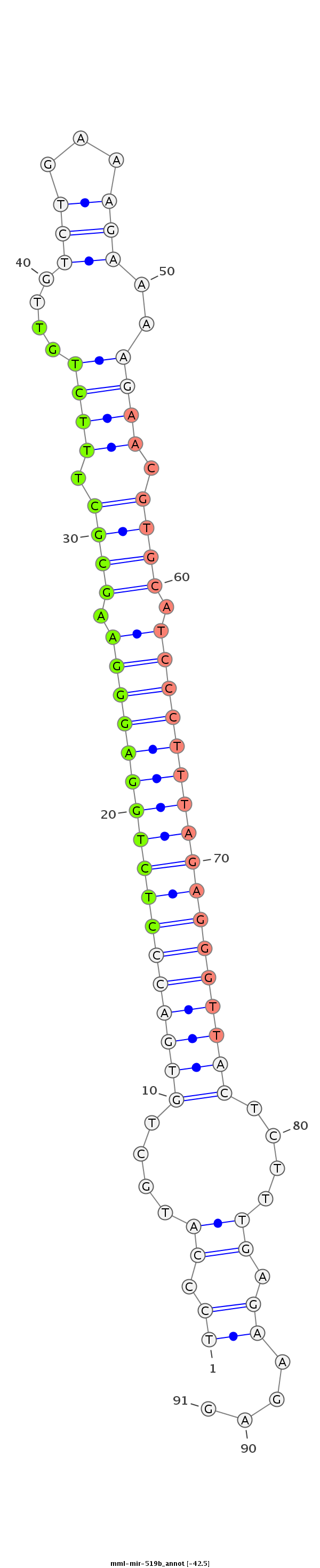
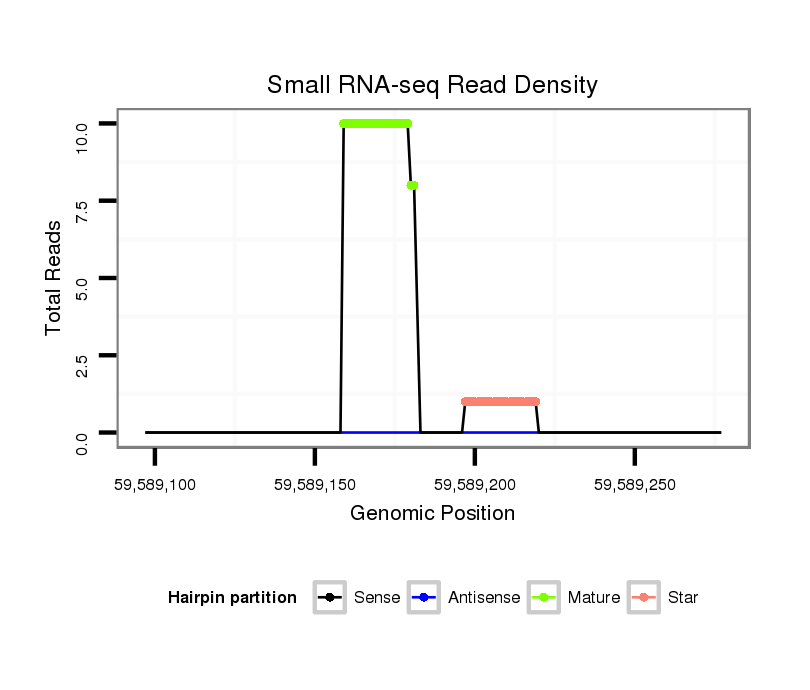
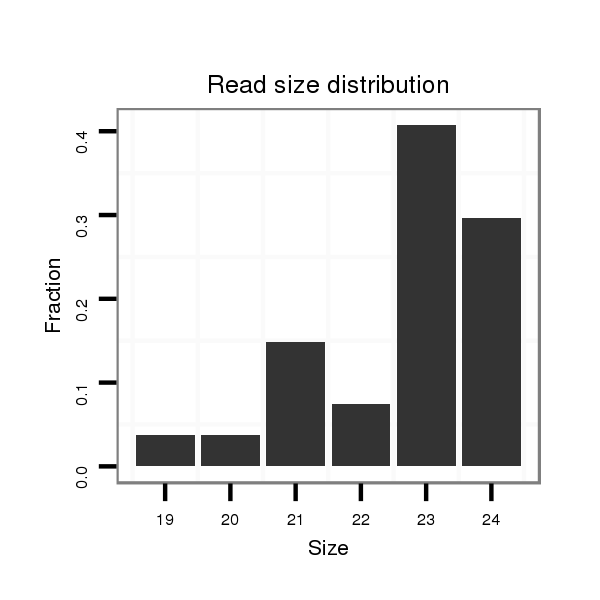
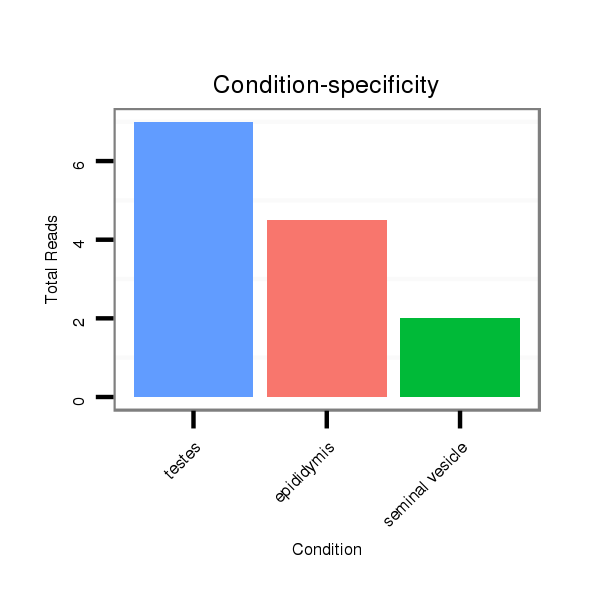
ID:mml-mir-519b |
Coordinate:chr19:59589147-59589227 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -41.6 |
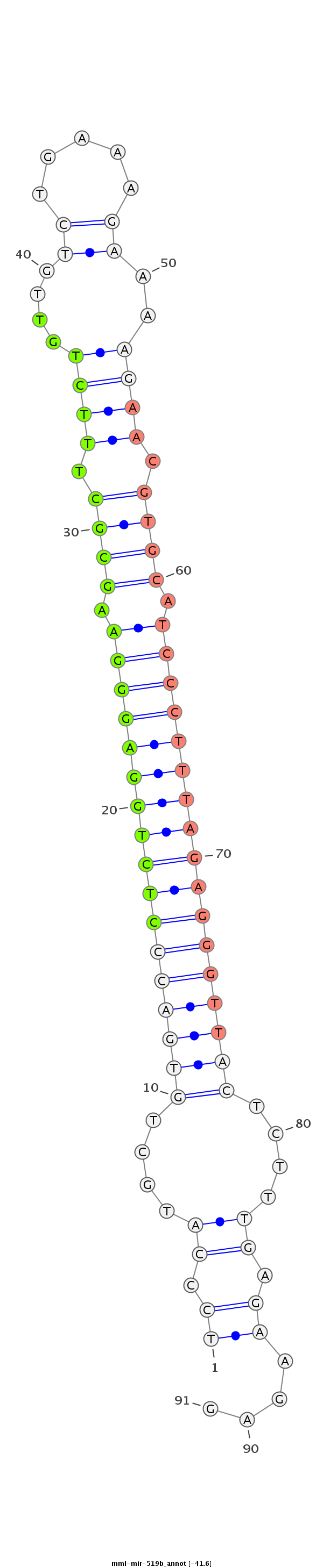 |
| mature | star |
|
GTCAAGGAAGATTCCAACAAAATATCCACGGTGCTGGAGCAAGAAGATCCCATGCTGTGACCCTCTGGAGGGAAGCGCTTTCTGTTGTCTGAAAGAAAAGAACGTGCATCCCTTTAGAGGGTTACTCTTTGAGAAGAGCAACGTTTAATTTGATGTTGATCTGGGTAATACGTTTGCAGAA
***********************************************((.((....(((((((((((((((((.((((.((((....(((...)))..)))).)))).)))))))))))))))))....)).))....******************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116839 testes |
SRR116840 epididymis |
SRR116842 seminal vesicle |
SRR116845 epididymis |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................CTCTGGAGGGAAGCGCTTTCTGT................................................................................................ | 23 | 0 | 2 | 4.50 | 9 | 3 | 3 | 3 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCTGTT............................................................................................... | 24 | 0 | 2 | 4.00 | 8 | 7 | 1 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCT.................................................................................................. | 21 | 0 | 2 | 2.00 | 4 | 2 | 2 | 0 | 0 |
| ....................................................................................................AACGTGCATCCCTTTAGAGGGTT.......................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTTTTTT............................................................................................... | 24 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCTG................................................................................................. | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCTAT................................................................................................ | 23 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................TCTGGAGGGAAGCGCTTTCTGT................................................................................................ | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCA.................................................................................................. | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................CTCTGGAGTGAAGCGCTTTCTGT................................................................................................ | 23 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTT.................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCTGTTT.............................................................................................. | 25 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCTGTTATC............................................................................................ | 27 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 |
| ..............................................................CTCTGGAGGGAAGCGCTTTT................................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTC................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTTCTTT................................................................................................ | 23 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................CTCTGGAGGGAAGCGCTTAA................................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .............................................................CCTCTCGAGGGAAGCGCTTTCT.................................................................................................. | 22 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
|
CAGTTCCTTCTAAGGTTGTTTTATAGGTGCCACGACCTCGTTCTTCTAGGGTACGACACTGGGAGACCTCCCTTCGCGAAAGACAACAGACTTTCTTTTCTTGCACGTAGGGAAATCTCCCAATGAGAAACTCTTCTCGTTGCAAATTAAACTACAACTAGACCCATTATGCAAACGTCTT
*******************************************((.((....(((((((((((((((((.((((.((((....(((...)))..)))).)))).)))))))))))))))))....)).))....*********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116844 epididymis |
|---|---|---|---|---|---|---|
| .........CTATGTTTGTTTTATAGGTGC....................................................................................................................................................... | 21 | 2 | 6 | 0.17 | 1 | 1 |
Generated: 09/01/2015 at 12:18 PM