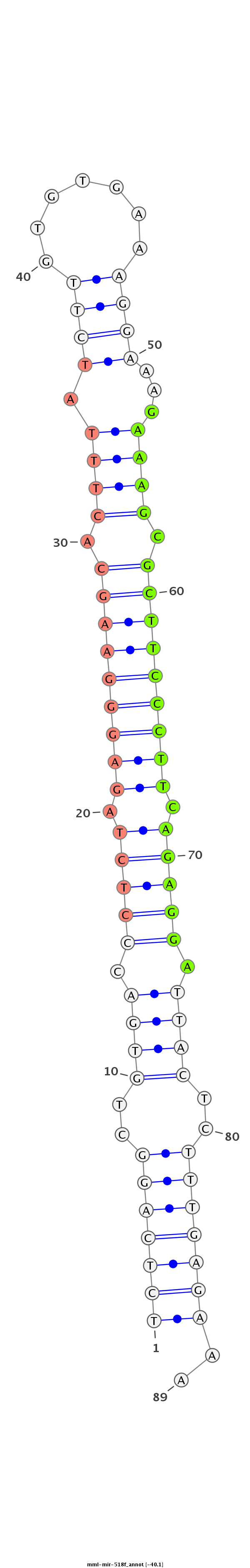

ID:mml-mir-518f |
Coordinate:chr19:59528995-59529081 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
CCAGTGAAGGAAAATTCCAACAAAAAATCCAGAGTGCTGGAGCAAGAAGATCTCAGGCTGTGACCCTCTAGAGGGAAGCACTTTATCTTGTGTGAAAGGAAAGAAAGCGCTTCCCTTCAGAGGATTACTCTTTGAGAAAAGCAACGTTGAAGTTGATGCTGATCTTGGTAATAGATTTGCAGAGAAT
**************************************************(((((((..((((.(((((.(((((((((.((((.((((.......))))...)))).))))))))).))))).))))..)))))))..************************************************ |
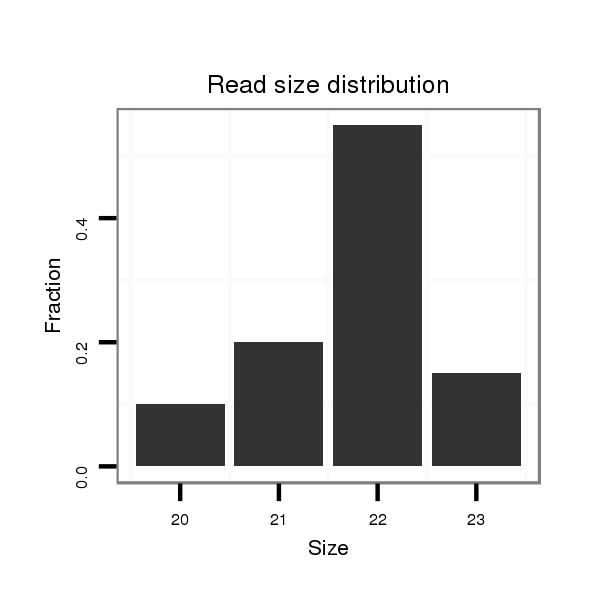
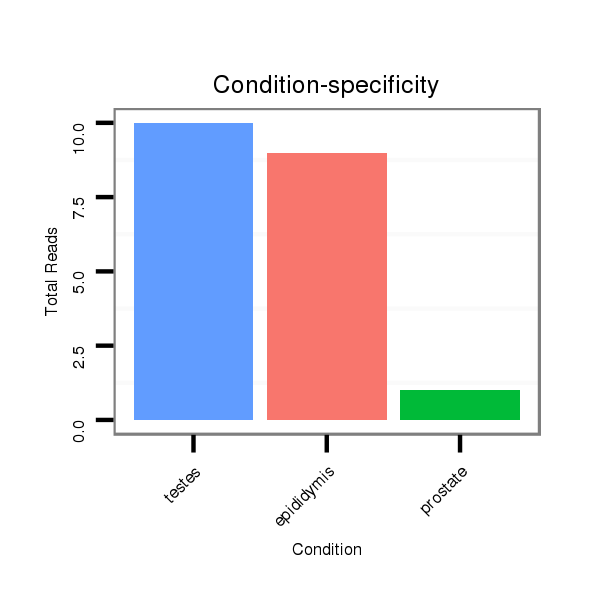
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116839 testes |
SRR116841 prostate |
SRR116842 seminal vesicle |
|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................GAAAGCGCTTCCCTTCAGAGGA............................................................... | 22 | 0 | 1 | 10.00 | 10 | 2 | 7 | 1 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTAT..................................................................................................... | 21 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 |
| ......................................................................................................GAAAGCGCTTCCCTTCAGAGGAT.............................................................. | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..................................................................TCTAGAGGGAAGCACTTTAT..................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................................GAAAGCGCTTCCCTTCAGAGGTTT............................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTATCT................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GAAAGCGCTTCCCTTCAGAGGATA............................................................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GAAAGCGCTTCCCTTCAGAGGAA.............................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GAAAGCGCTTCCCTTCCGAGGA............................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTATC.................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GAAAGAGCTTCCCTTCAGAGGA............................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ................................................................CCTCTAGAAGGAAGCACTTTA...................................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................CCCTCTAGAAGGAAGCACTTTAA..................................................................................................... | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................CTCTAGAGGGAAGCACTTTT...................................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................GAAAGCGCTTCCCTTCAGT.................................................................. | 19 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 |
| .................................................................GTCTAGAGGGAAGCACTTTAA..................................................................................................... | 21 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ............................................AGAAGATCTCAGGCTGTGAC........................................................................................................................... | 20 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 |
|
GGTCACTTCCTTTTAAGGTTGTTTTTTAGGTCTCACGACCTCGTTCTTCTAGAGTCCGACACTGGGAGATCTCCCTTCGTGAAATAGAACACACTTTCCTTTCTTTCGCGAAGGGAAGTCTCCTAATGAGAAACTCTTTTCGTTGCAACTTCAACTACGACTAGAACCATTATCTAAACGTCTCTTA
************************************************(((((((..((((.(((((.(((((((((.((((.((((.......))))...)))).))))))))).))))).))))..)))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116839 testes |
SRR116840 epididymis |
|---|---|---|---|---|---|---|---|
| ...................................................................................................TTTCTTTCGCGAAGGGAGGTCTC................................................................. | 23 | 1 | 3 | 0.33 | 1 | 1 | 0 |
| ...................................................................................................TTTCTTTCGCGAAGGGAAATCTC................................................................. | 23 | 1 | 4 | 0.25 | 1 | 0 | 1 |
Generated: 09/01/2015 at 12:16 PM