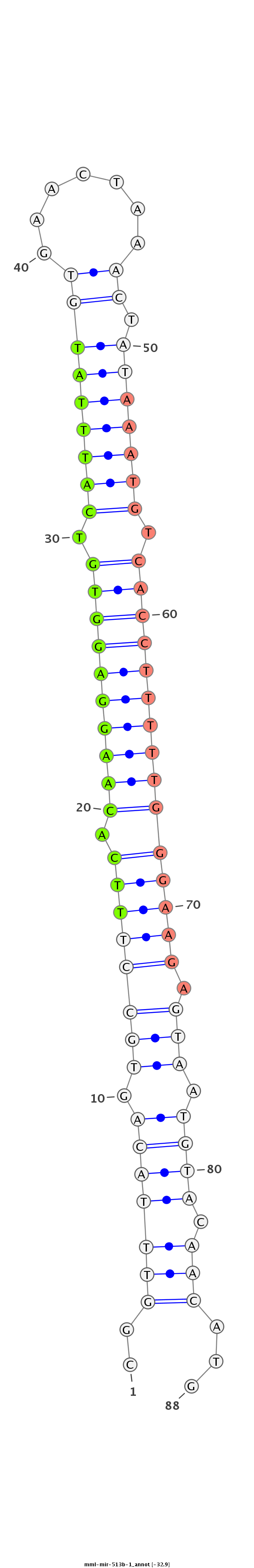
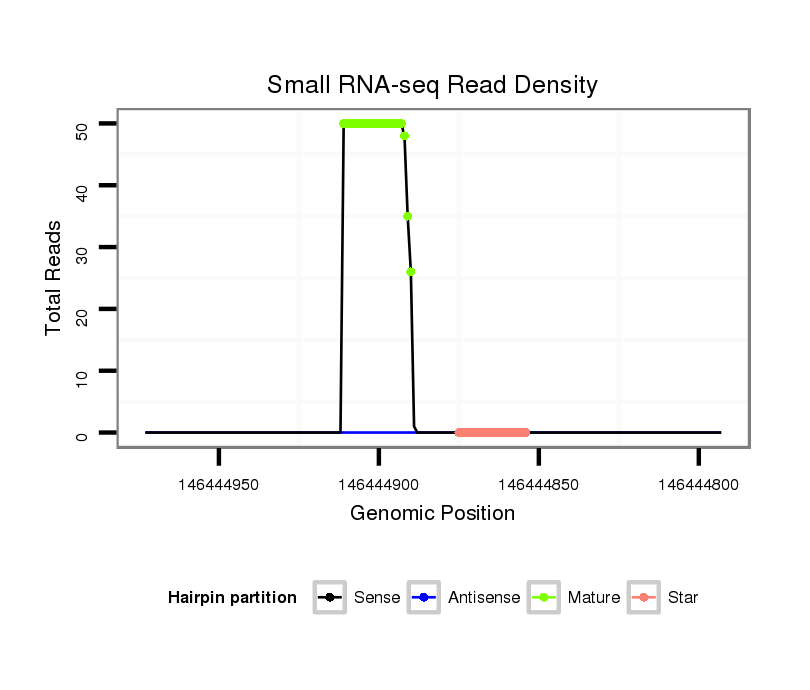
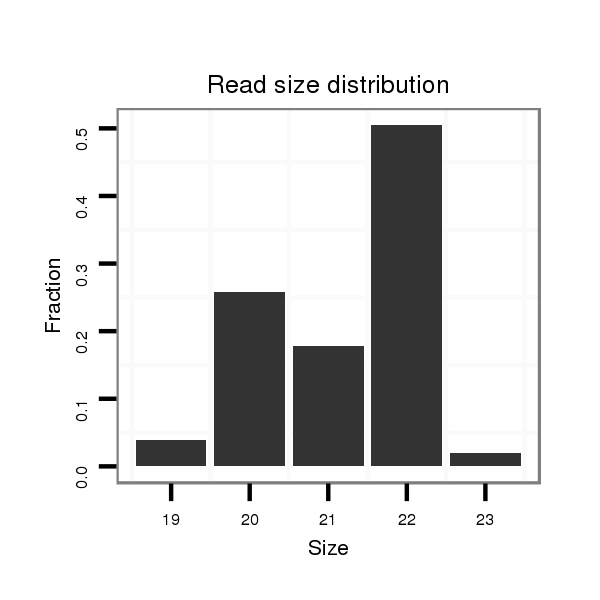
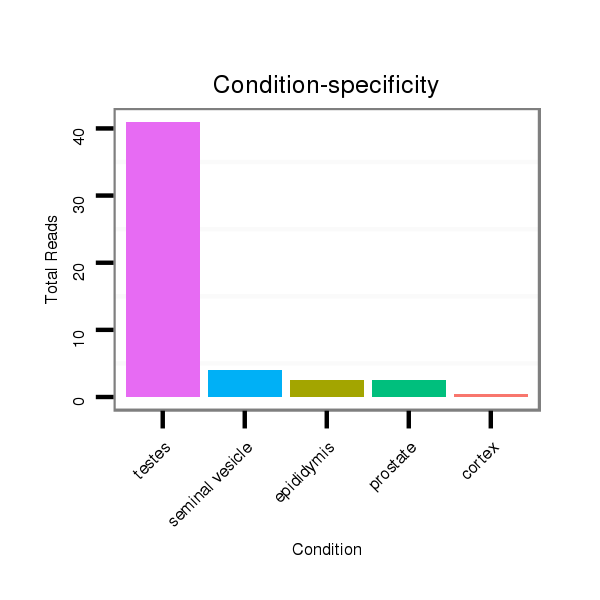
ID:mml-mir-513b-1 |
Coordinate:chrX:146444843-146444923 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -32.3 |
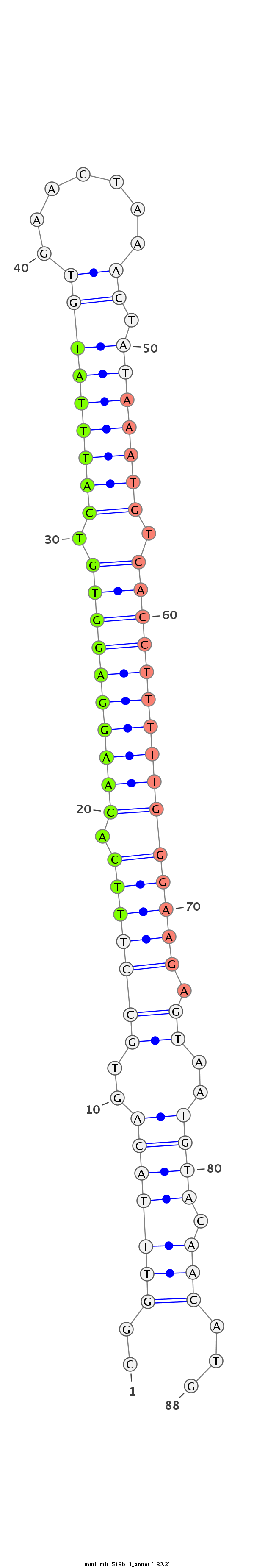 |
| mature | star |
|
TGGCATGAATAGGGAGCATTGGGTCTGGGATGCCACATTCAGCCATTCGGTTTACAGTGCCTTTCACAAGGAGGTGTCATTTATGTGAACTAAACTATAAATGTCACCTTTTTGGGAAGAGTAATGTACAACATGCACTGCAAATGTGGTGTCCCTGGGGATGGGGCTGTAGCTGCGAGCA
***********************************************..(((((((.((((((((.((((((((((.(((((((((.......)).))))))).))))))))))))))).))).)))).)))...********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116839 testes |
SRR116842 seminal vesicle |
SRR116841 prostate |
SRR116840 epididymis |
SRR116843 cortex |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................TTCACAAGGAGGTGTCATTTAT................................................................................................. | 22 | 0 | 2 | 25.00 | 50 | 41 | 2 | 4 | 2 | 1 |
| ..............................................................TTCACAAGGAGGTGTCATTT................................................................................................... | 20 | 0 | 2 | 13.00 | 26 | 21 | 3 | 0 | 2 | 0 |
| ..............................................................TTCACAAGGAGGTGTCATTTA.................................................................................................. | 21 | 0 | 2 | 9.00 | 18 | 15 | 2 | 1 | 0 | 0 |
| ..............................................................TTCACAAGGAGGTGTCATT.................................................................................................... | 19 | 0 | 2 | 2.00 | 4 | 2 | 1 | 0 | 1 | 0 |
| ..............................................................TTCACAAGGAGGTGTCATTTATG................................................................................................ | 23 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACAAGGAGGTGTCATTTAA................................................................................................. | 22 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACAAGGAGGTGTCATTTATT................................................................................................ | 23 | 1 | 2 | 1.00 | 2 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................TTCACGAGGAGGTGTCATTTAT................................................................................................. | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACCAGGAGGTGTCATTTAT................................................................................................. | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACAAGGAGGGGTCATTTAT................................................................................................. | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................TTCACAAGGAGTTGTCATTTAT................................................................................................. | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACCAGGAGGTGTCATTTA.................................................................................................. | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACAAGGAGGTGTCATTAA.................................................................................................. | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TTCGCAAGGAGGTGTCATTTAT................................................................................................. | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACAAGGAGGTGTTATTTAT................................................................................................. | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................AAATGTCACCTTTTTGGGAAGA............................................................. | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................TTCACTAGGAGGTGTCATTTA.................................................................................................. | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
|
ACCGTACTTATCCCTCGTAACCCAGACCCTACGGTGTAAGTCGGTAAGCCAAATGTCACGGAAAGTGTTCCTCCACAGTAAATACACTTGATTTGATATTTACAGTGGAAAAACCCTTCTCATTACATGTTGTACGTGACGTTTACACCACAGGGACCCCTACCCCGACATCGACGCTCGT
**********************************************..(((((((.((((((((.((((((((((.(((((((((.......)).))))))).))))))))))))))).))).)))).)))...*********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:37 PM