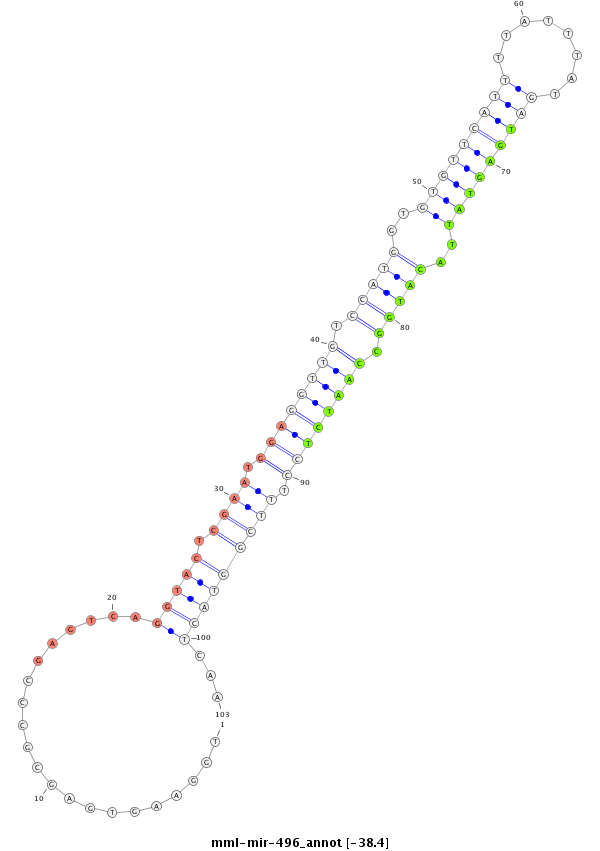
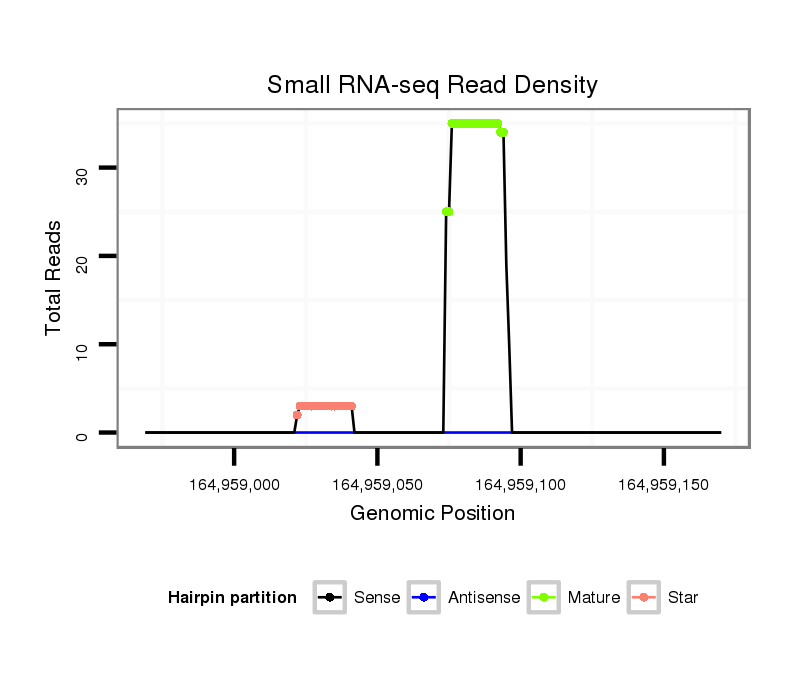
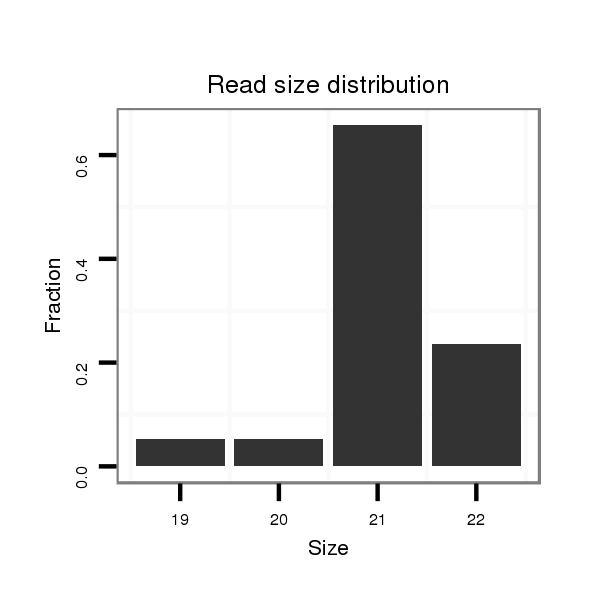
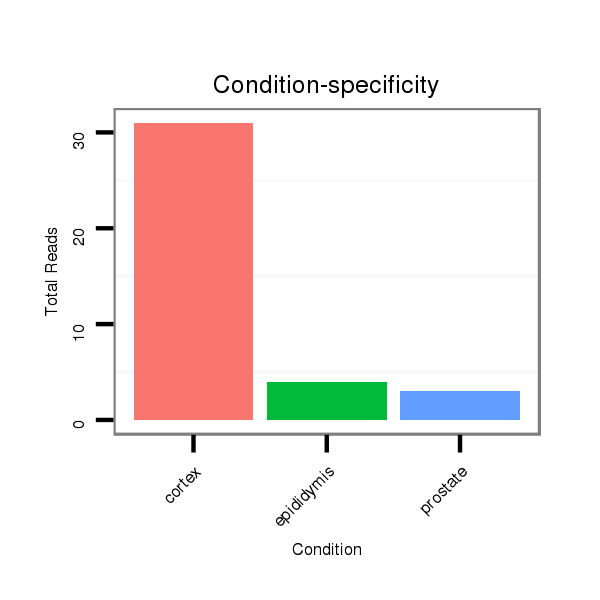
ID:mml-mir-496 |
Coordinate:chr7:164959019-164959120 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -38.3 | -38.3 | -38.3 |
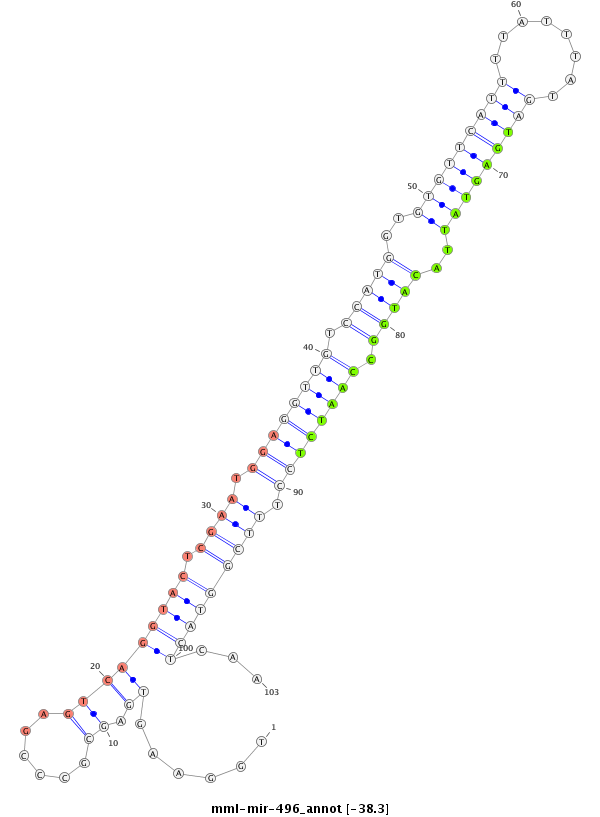 |
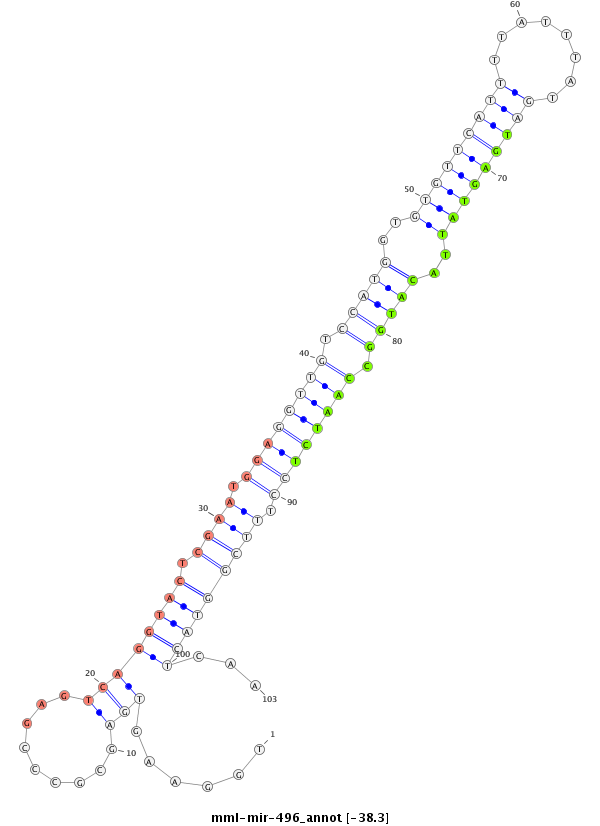 |
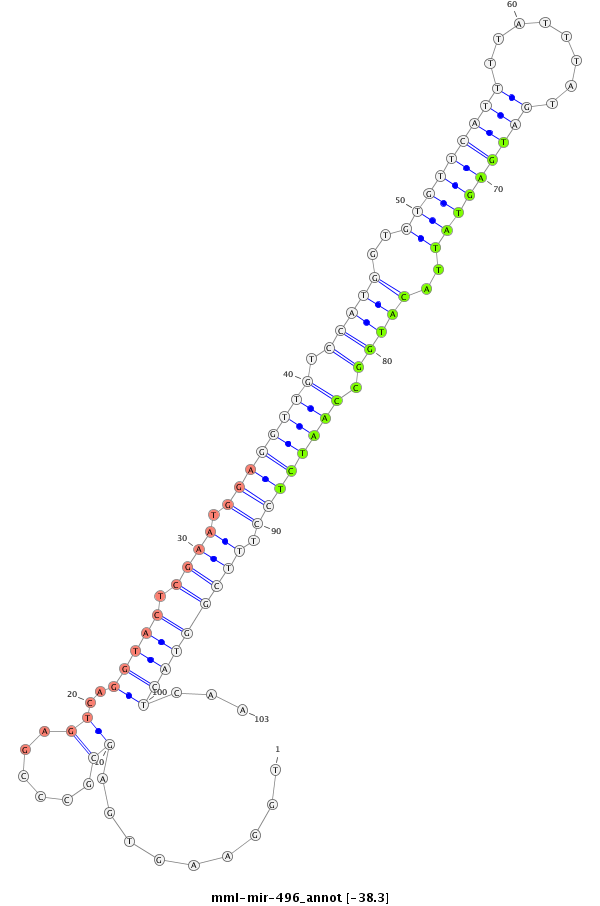 |
|
GAATCCCGTCCGTCCTCAGGCCTGCTGCTGGGGCGCGCTGGAAGTGAGCGCCCGAGTCAGGTACTCGAATGGAGGTTGTCCATGGTGTGTTCATTTTATTTATGATGAGTATTACATGGCCAATCTCCTTTCGGTACTCAATTCTTCTTGGGAAACGTCACGAGGGGCAAGCCCAGCCGGCACCTGCTCAGGGTAAGGAAGT
**************************************.....................(((((.((((.((((((((.(((((..(((((((((........)))))))))..))))).)))))))).)))))))))...************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116840 epididymis |
SRR116841 prostate |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TGAGTATTACATGGCCAATCT............................................................................ | 21 | 0 | 1 | 15.00 | 15 | 12 | 2 | 1 | 0 |
| ...........................................................................................................AGTATTACATGGCCAATCTCC.......................................................................... | 21 | 0 | 1 | 10.00 | 10 | 8 | 1 | 1 | 0 |
| .........................................................................................................TGAGTATTACATGGCCAATCTC........................................................................... | 22 | 0 | 1 | 9.00 | 9 | 7 | 1 | 1 | 0 |
| .....................................................GAGTCAGGTACTCGAATGGA................................................................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................................AGTATTACATGGCCAATCTCT.......................................................................... | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................................AGTATTACATGGCCAATCTCCATC....................................................................... | 24 | 2 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 |
| .........................................................................................................TGAGTATTACATGGCCCATCT............................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................TGCGTATTACATGGCCCATCTC........................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................TGAGTATTACATTGCCAATCTC........................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................AGTATTACATGGCCAATCTCCAA........................................................................ | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................TGAGTATTACATGTCCAATCTC........................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................TGAGTATTACATGGCCAAT.............................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................AGTCAGGTACTCGAATGGA................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................TGAGTATTACATGGCCAATCTATC......................................................................... | 24 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 2 |
| ........................CTGCGGGGGCGCGCTGG................................................................................................................................................................. | 17 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
|
CTTAGGGCAGGCAGGAGTCCGGACGACGACCCCGCGCGACCTTCACTCGCGGGCTCAGTCCATGAGCTTACCTCCAACAGGTACCACACAAGTAAAATAAATACTACTCATAATGTACCGGTTAGAGGAAAGCCATGAGTTAAGAAGAACCCTTTGCAGTGCTCCCCGTTCGGGTCGGCCGTGGACGAGTCCCATTCCTTCA
*************************************************************.....................(((((.((((.((((((((.(((((..(((((((((........)))))))))..))))).)))))))).)))))))))...************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:31 PM