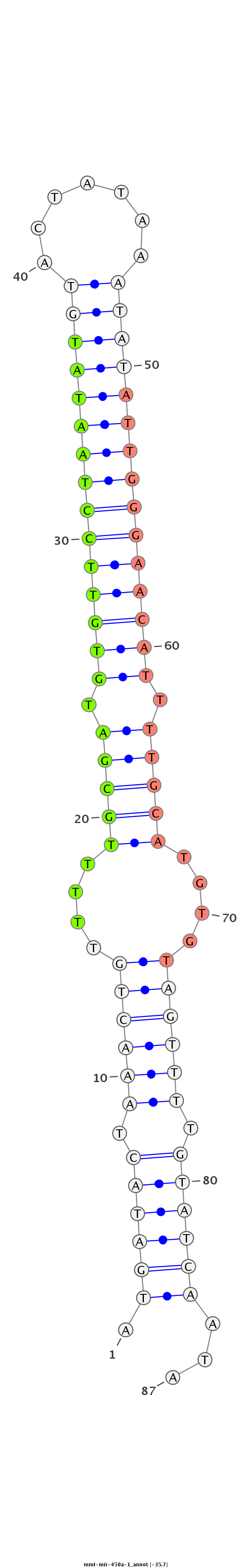



ID:mml-mir-450a-1 |
Coordinate:chrX:133862543-133862633 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
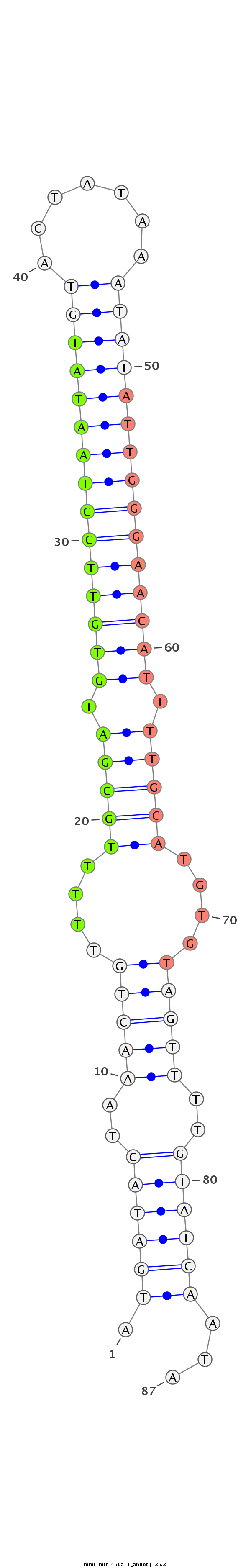
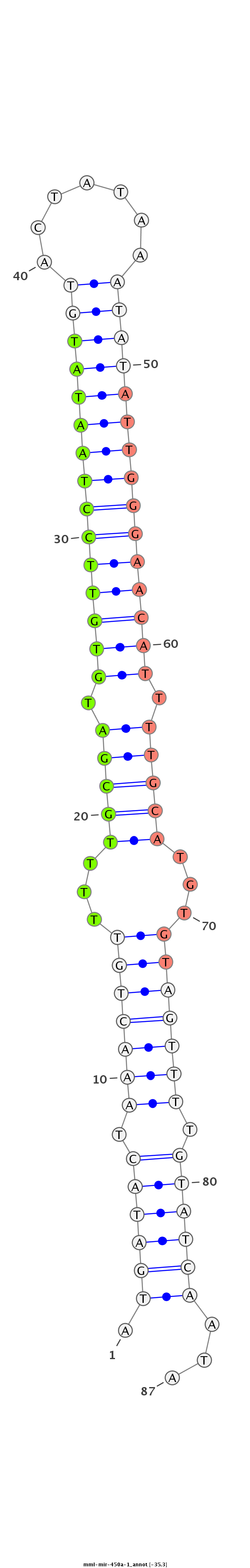
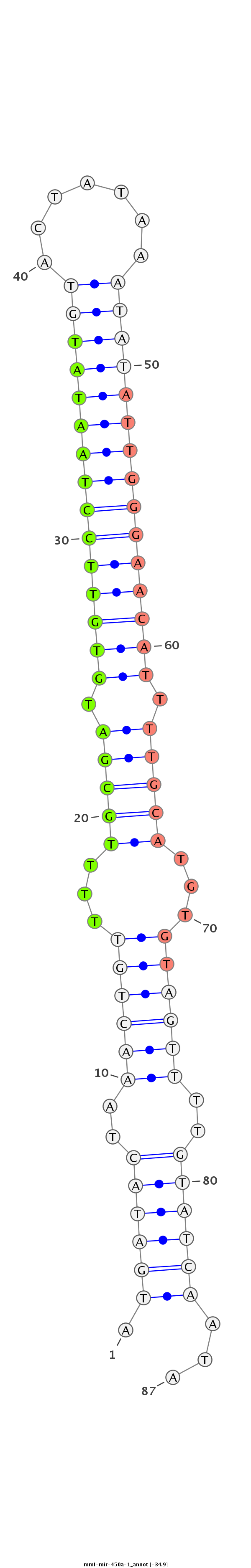
| -35.3 | -35.3 | -34.9 |
 |
 |
 |
| mature | star |
|
CAAAAGTCTAGAGGTGGTTATCGCTGACTTGTGTCACTATAGGCAATCAAAAATGATACTAAACTGTTTTTGCGATGTGTTCCTAATATGTACTATAAATATATTGGGAACATTTTGCATGTGTAGTTTTGTATCAATATACAGATGAAACGATGGAGGAAATAAACCAAAAATCTAACAGTGGCTATCTG
****************************************************.((((((.((((((....(((((.(((((((((((((((.......))))))))))))))).)))))....)))))).))))))...**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116845 epididymis |
SRR116844 epididymis |
SRR116842 seminal vesicle |
SRR116840 epididymis |
SRR116841 prostate |
SRR116839 testes |
SRR116843 cortex |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................TTTTGCGATGTGTTCCTAATAATC.................................................................................................... | 24 | 3 | 2 | 7.50 | 15 | 7 | 8 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAATAT...................................................................................................... | 22 | 0 | 2 | 7.50 | 15 | 0 | 0 | 4 | 6 | 3 | 1 | 1 |
| ...................................................................TTTTGCGATGTGTTCCTAATATATC................................................................................................... | 25 | 2 | 2 | 7.00 | 14 | 9 | 5 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAAT........................................................................................................ | 20 | 0 | 2 | 6.50 | 13 | 0 | 0 | 6 | 4 | 2 | 1 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAATATC..................................................................................................... | 23 | 1 | 2 | 5.50 | 11 | 2 | 9 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ATTGGGAACATTTTGCATGTGTATC................................................................ | 25 | 2 | 1 | 4.00 | 4 | 1 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAATA....................................................................................................... | 21 | 0 | 2 | 3.50 | 7 | 0 | 0 | 2 | 1 | 1 | 2 | 1 |
| ...................................................................TTTTGCGATGTGTTCCTAATATGATC.................................................................................................. | 26 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAATATTATC.................................................................................................. | 26 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TTGGGAACATTTTGCATGTGTATCC............................................................... | 25 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAA......................................................................................................... | 19 | 0 | 2 | 1.50 | 3 | 0 | 0 | 1 | 0 | 1 | 1 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAATATAT.................................................................................................... | 24 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ATTGGGAACATTTTGCATGTGA................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................ATTGGGAACATTTTGCATGTGT................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCC............................................................................................................ | 16 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCCAAT........................................................................................................ | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTTTTCCTAATAT...................................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAATAA...................................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................ATTGGGAACATTTTGCATGTGATC................................................................. | 24 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGAGATGTGTTCCTAATAT...................................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................TTTGCGATGTGTTCCTAAT........................................................................................................ | 19 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTA.......................................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCCTAATATA..................................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................TTTTGCGATGTGTTCATAATATATC................................................................................................... | 25 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TTGGGAACATTTTGCATGTGATC................................................................. | 23 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTTTTCAGATCTCCACCAATAGCGACTGAACACAGTGATATCCGTTAGTTTTTACTATGATTTGACAAAAACGCTACACAAGGATTATACATGATATTTATATAACCCTTGTAAAACGTACACATCAAAACATAGTTATATGTCTACTTTGCTACCTCCTTTATTTGGTTTTTAGATTGTCACCGATAGAC
****************************************************.((((((.((((((....(((((.(((((((((((((((.......))))))))))))))).)))))....)))))).))))))...**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:36 PM