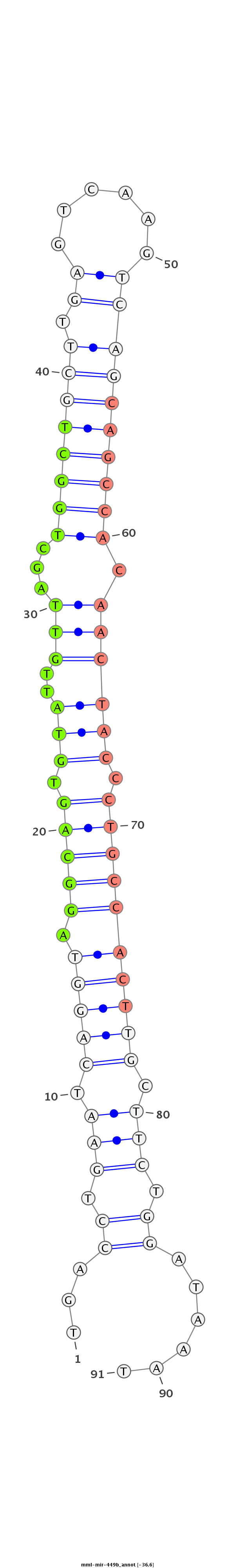
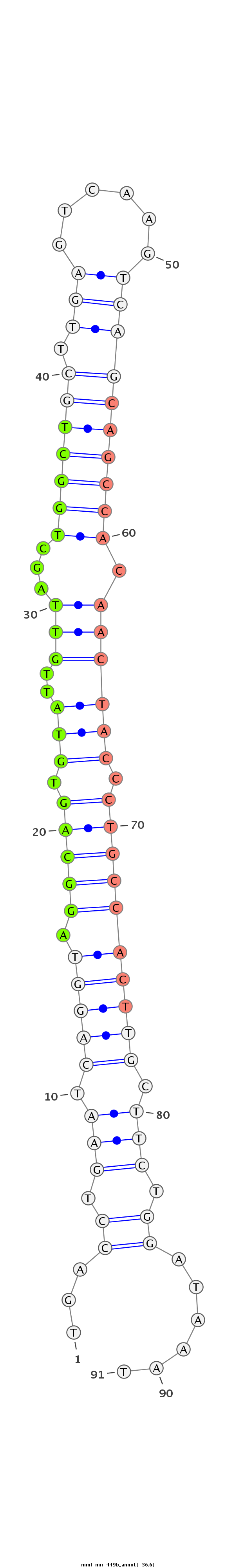
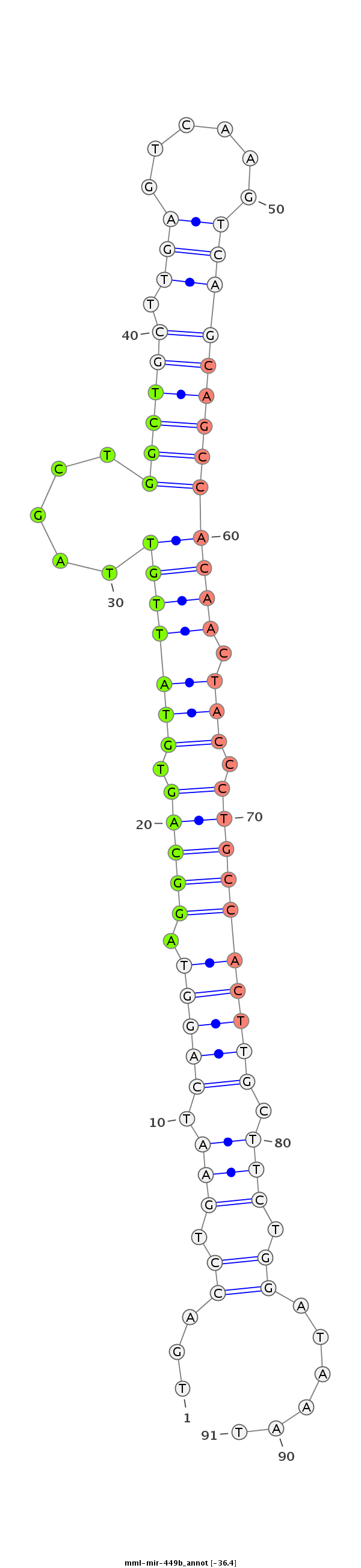
ID:mml-mir-449b |
Coordinate:chr6:53216425-53216521 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -36.6 | -36.4 | -36.4 |
 |
 |
 |
|
CATGTTACAAATTTAGCCTCGGTGACTTTCTGGGCCACGAGAATCAGCAGTGACCTGAATCAGGTAGGCAGTGTATTGTTAGCTGGCTGCTTGAGTCAAGTCAGCAGCCACAACTACCCTGCCACTTGCTTCTGGATAAATTCTTCTTGTCAATGAAGTGCTCTGGCTACCTGTGTGTGATGAGCTGGCAGTGTATT
**************************************************...((.(((.(((((.(((((.(((..(((...((((((((.((......)))))))))).)))))).)))))))))).))).))......******************************************************** |
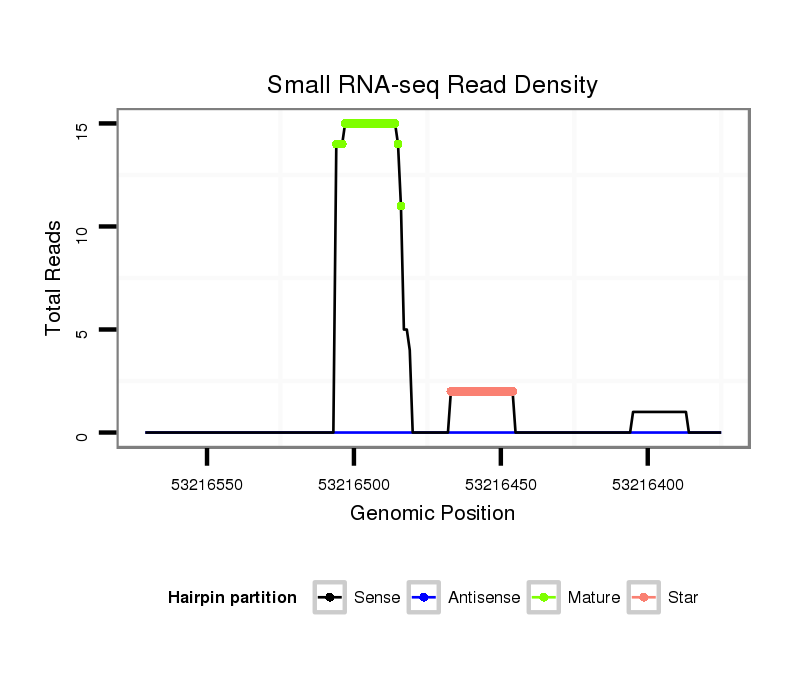
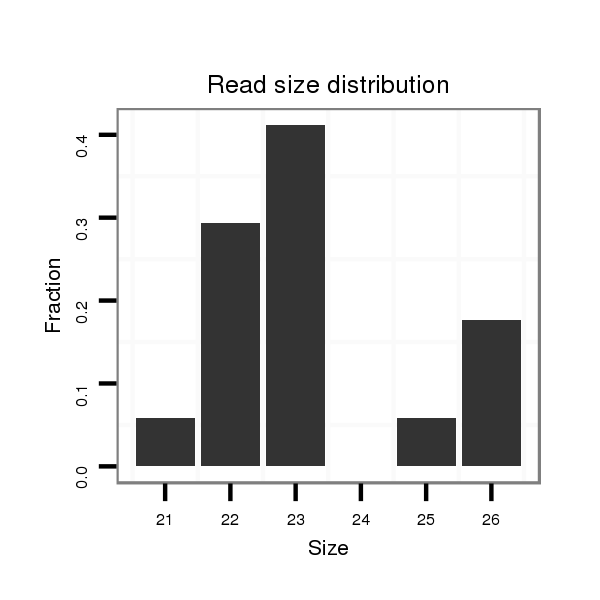
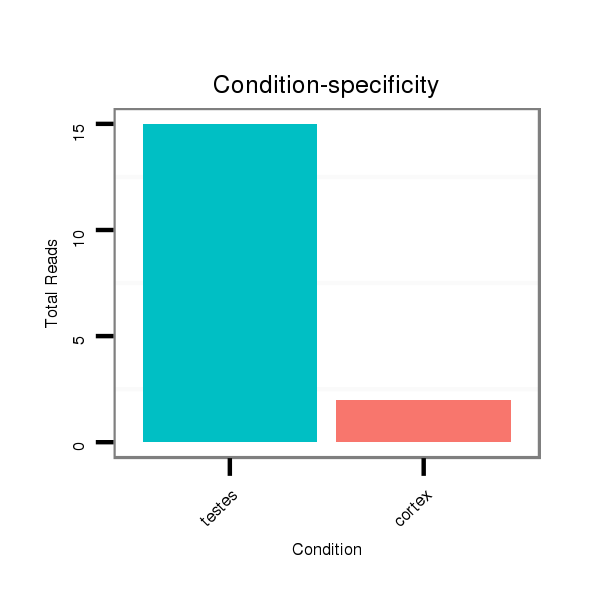
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116839 testes |
SRR116843 cortex |
SRR116840 epididymis |
|---|---|---|---|---|---|---|---|---|
| .................................................................AGGCAGTGTATTGTTAGCTGGCT............................................................................................................. | 23 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGC.............................................................................................................. | 22 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGCTGCT.......................................................................................................... | 26 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTTGCT............................................................................................................. | 23 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 |
| ........................................................................................................CAGCCACAACTACCCTGCCACT....................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGCTGC........................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGCTA............................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGCTTT........................................................................................................... | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................................................CTACCTGTGTGTGATGAGC............ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTTGCTGC........................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGG............................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .................................................................AGGCAGTGTATTTTTAGCTGGCTTT........................................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGCTAA........................................................................................................... | 25 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGAA............................................................................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTTGCTGCAT......................................................................................................... | 27 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ....................................................................CAGTGTATTGTTAGCTGGCTGCT.......................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGTTAGCTGGCA............................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................AGGCAGTGTATTGGTAGCTGGG.............................................................................................................. | 22 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 |
| .................................................................AGGCAGTGTATTTTTAGCTGGCA............................................................................................................. | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ........................................................................................................CAGCCACAACTACCCTGCCACAAA..................................................................... | 24 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 |
|
GTACAATGTTTAAATCGGAGCCACTGAAAGACCCGGTGCTCTTAGTCGTCACTGGACTTAGTCCATCCGTCACATAACAATCGACCGACGAACTCAGTTCAGTCGTCGGTGTTGATGGGACGGTGAACGAAGACCTATTTAAGAAGAACAGTTACTTCACGAGACCGATGGACACACACTACTCGACCGTCACATAA
********************************************************...((.(((.(((((.(((((.(((..(((...((((((((.((......)))))))))).)))))).)))))))))).))).))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:25 PM