
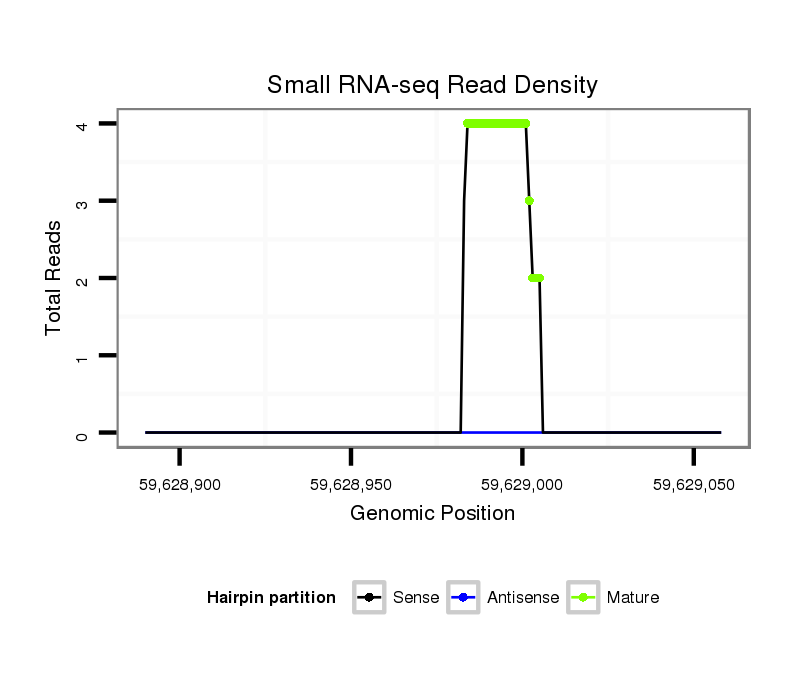

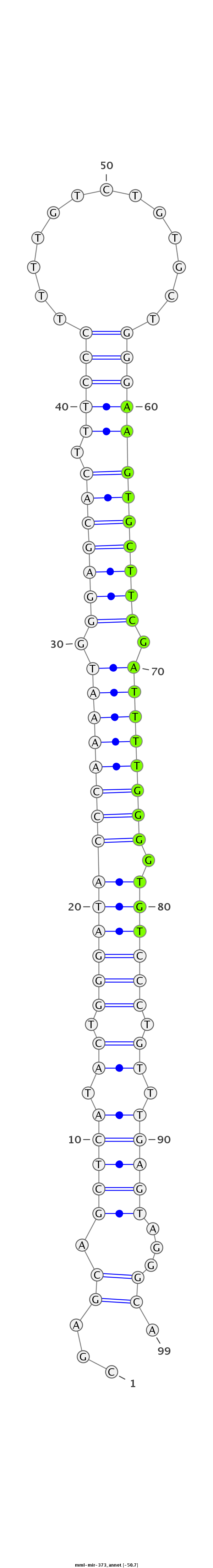
ID:mml-mir-373 |
Coordinate:chr19:59628940-59629008 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -50.7 | -50.7 | -50.7 |
 |
 |
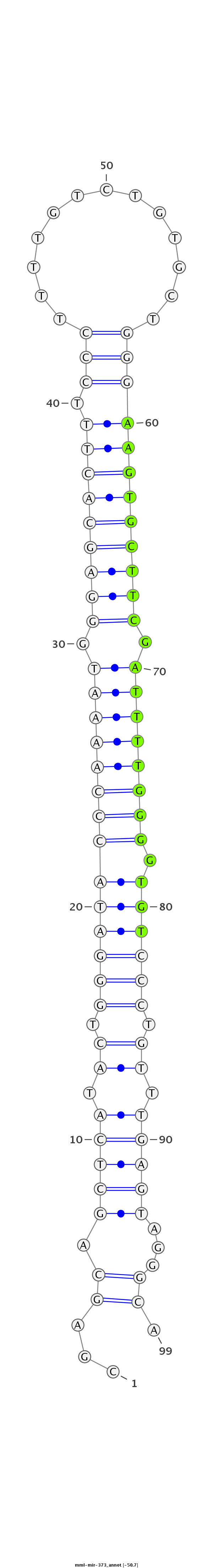 |
|
GTAGGCAAGAAAGTCGCAGTGATGGCAGATCCTTGCGAGCAGCTCATACTGGGATACCCAAAATGGGAGCACTTTCCCTTTTGTCTGTGCTGGGAAGTGCTTCGATTTTGGGGTGTCCCTGTTTGAGTAGGGCATCACAAACCATCCTGCTTCAAGGGAGCCTGCGGGT
***********************************...((.(((((.((.((((((((((((((.(((((((((.(((.............)))))))))))).))))).))))))))).)).)))))...)).*********************************** |
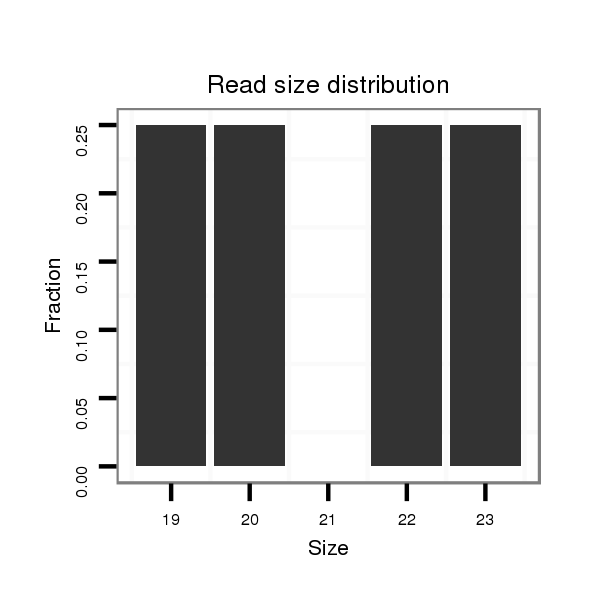
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116845 epididymis |
SRR116844 epididymis |
SRR116840 epididymis |
SRR116839 testes |
SRR116841 prostate |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGTATC.................................................. | 26 | 2 | 1 | 175.00 | 175 | 147 | 28 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGATC................................................... | 25 | 2 | 1 | 33.00 | 33 | 27 | 6 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGAATC.................................................. | 26 | 3 | 1 | 26.00 | 26 | 26 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTATC.................................................... | 24 | 1 | 1 | 23.00 | 23 | 23 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGTTGTATC.................................................. | 26 | 3 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGTA.................................................... | 24 | 1 | 1 | 4.00 | 4 | 3 | 0 | 0 | 1 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTTGGGTGTATC.................................................. | 26 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTAATC................................................... | 25 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGGGTATC.................................................. | 26 | 3 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGTAT................................................... | 25 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGATC..................................................... | 23 | 3 | 3 | 2.67 | 8 | 7 | 1 | 0 | 0 | 0 |
| ..............................................................................................AAGTGCTTCGATTTTGGGGTGTAGAT................................................. | 26 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .............................................................................................GACGTGCTTCGATTTTGGGGTGTATC.................................................. | 26 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTTGGGTGTTC................................................... | 25 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTCCGATTTTGGGGTGTATC.................................................. | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................AAGTGCTTCGATTTTGGGGTGT..................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTTTATC.................................................. | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGAATC.................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGTGGTGTATC.................................................. | 26 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGG........................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................GTGCTTCGATTTTGGGGTATC.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGG......................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGCTTTTGGGGTGTATC.................................................. | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTATATC.................................................. | 26 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGT..................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................AAGTGCTTCGATTTTGGGGTGA..................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGCGTATC.................................................. | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTTGGGTATC.................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTCGGGTGATC................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGCATC.................................................. | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGAAC................................................... | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGAGTGTATC.................................................. | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGGGTGAT.................................................... | 24 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................GAAGTGCTTCGATTTTGGAATC...................................................... | 22 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
|
CATCCGTTCTTTCAGCGTCACTACCGTCTAGGAACGCTCGTCGAGTATGACCCTATGGGTTTTACCCTCGTGAAAGGGAAAACAGACACGACCCTTCACGAAGCTAAAACCCCACAGGGACAAACTCATCCCGTAGTGTTTGGTAGGACGAAGTTCCCTCGGACGCCCA
***********************************...((.(((((.((.((((((((((((((.(((((((((.(((.............)))))))))))).))))).))))))))).)).)))))...)).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116842 seminal vesicle |
|---|---|---|---|---|---|---|
| ................................................................................................................................TCCCGTAGTGTTTGGA......................... | 16 | 1 | 20 | 0.05 | 1 | 1 |
Generated: 09/01/2015 at 12:18 PM