
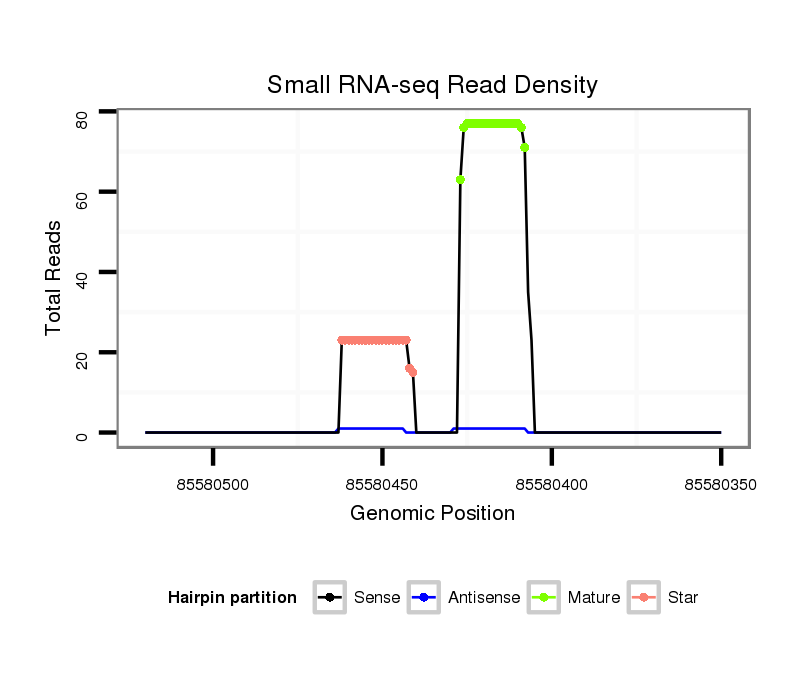
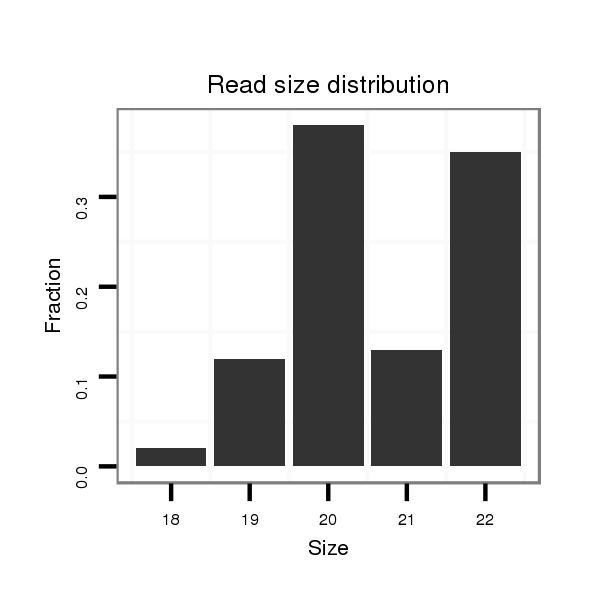
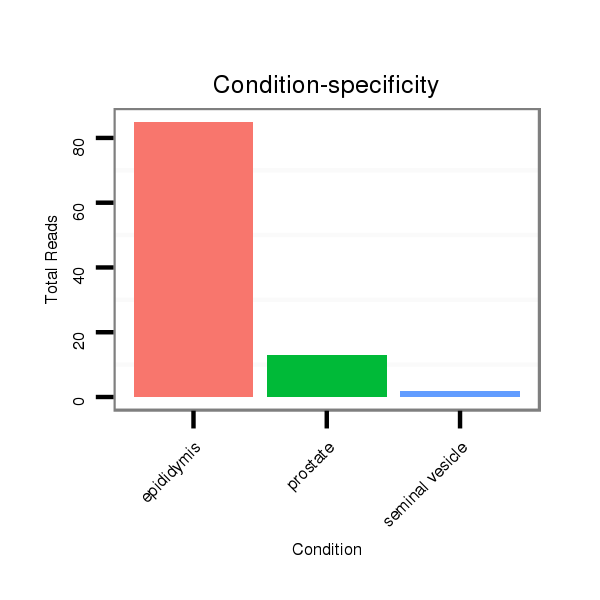
ID:mml-mir-208a |
Coordinate:chr7:85580400-85580470 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -39.2 | -39.2 | -39.0 |
 |
 |
 |
|
GCCTGAACTCTATACTCTGTGAACTCTGGCTGGCCCTGACCCGCTTCCTGTGACAGGCGAGCTTTTGGCCCGGGTTATACCTGATGCTCACGTATAAGACGAGCAAAAAGCTTGTTGGTCAGAGGAGCTACCGTCGATCAGCCTGTGTGGGGGGCGAGGGCAGGGGGCACT
*******************************************.(((((.((((.(((((((((((.((.((..((((((.(((...))).))))))..)).)).))))))))))).)))).))))).******************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116841 prostate |
SRR116842 seminal vesicle |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................ATAAGACGAGCAAAAAGCTT.......................................................... | 20 | 0 | 1 | 29.00 | 29 | 28 | 1 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGCTTGT........................................................ | 22 | 0 | 1 | 20.00 | 20 | 15 | 5 | 0 | 0 |
| ..........................................................GAGCTTTTGGCCCGGGTTATAC........................................................................................... | 22 | 0 | 1 | 15.00 | 15 | 14 | 1 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGCTTG......................................................... | 21 | 0 | 1 | 9.00 | 9 | 7 | 1 | 1 | 0 |
| ..........................................................GAGCTTTTGGCCCGGGTTAT............................................................................................. | 20 | 0 | 1 | 7.00 | 7 | 6 | 1 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCTT.......................................................... | 19 | 0 | 1 | 7.00 | 7 | 5 | 2 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCTTGTAA...................................................... | 23 | 2 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGCT........................................................... | 19 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCTTGTTT...................................................... | 23 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCTTGT........................................................ | 21 | 0 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 |
| .............................................................................................ATAAGACGAGTAAAAAGCTT.......................................................... | 20 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCTTG......................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..........................................................GAGCTTTTGGCCCGGGTTATACT.......................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 |
| .............................................................................................ATAAGACGGGCAAAAAGCTTG......................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................AAGACGAGCAAAAAGCTTG......................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................GAGCTTTTGGCCCGGGTTATAT........................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................ATAAGACGACCAAAAAGCTTAT........................................................ | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGCTTA......................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGC............................................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................GAGCTTTTTGCCCGGGTTAT............................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................GAGCTTTTGGCCCGGGTTATACA.......................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCTTGTTA...................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................AAGACGAGCAAAAAGCTTGTAA...................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCT........................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................ATAGGACGAGCAAAAAGCTTGT........................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................ATAAGGCGAGCAAAAAGCTTG......................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................GAGCTTTTGGCCCGGGTTATA............................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGCTTT......................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................AAGACGAGCAAAAAGCTTTTT....................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................ATCAGACGAGCAAAAAGCTTG......................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................ATCAGACGAGCAAAAAGCTTGT........................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................AAGACGAGCAAAAAGCTTGTTA...................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCTTTTT....................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................GAGCTTTTGGCCCGGGTTATACAC......................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..............................................................................................TAAGACGAGCAAAAAGCTTGTA....................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................TAAGACGAGCAAAAAGCA........................................................... | 18 | 1 | 5 | 0.80 | 4 | 4 | 0 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGCA........................................................... | 19 | 1 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 |
| .............................................................................................ATAAGACGAGCAAAAAGCTA.......................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................GAGCTTTTGTCCCGTGTTATAC........................................................................................... | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................ATAATACGAGCAAAAAGCTT.......................................................... | 20 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
|
CGGACTTGAGATATGAGACACTTGAGACCGACCGGGACTGGGCGAAGGACACTGTCCGCTCGAAAACCGGGCCCAATATGGACTACGAGTGCATATTCTGCTCGTTTTTCGAACAACCAGTCTCCTCGATGGCAGCTAGTCGGACACACCCCCCGCTCCCGTCCCCCGTGA
*******************************************.(((((.((((.(((((((((((.((.((..((((((.(((...))).))))))..)).)).))))))))))).)))).))))).******************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116841 prostate |
SRR116844 epididymis |
SRR116845 epididymis |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................TATTCTTCTCGTTTTTCGAA.......................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................GCTCGAAAACCGGGCCCAAT.............................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................CATATTCTGCTCGTTTTTCGAA.......................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................CTACGCGTGCATAATCTG....................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................CAAGTCTCCTCGATG........................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 09/01/2015 at 12:27 PM