
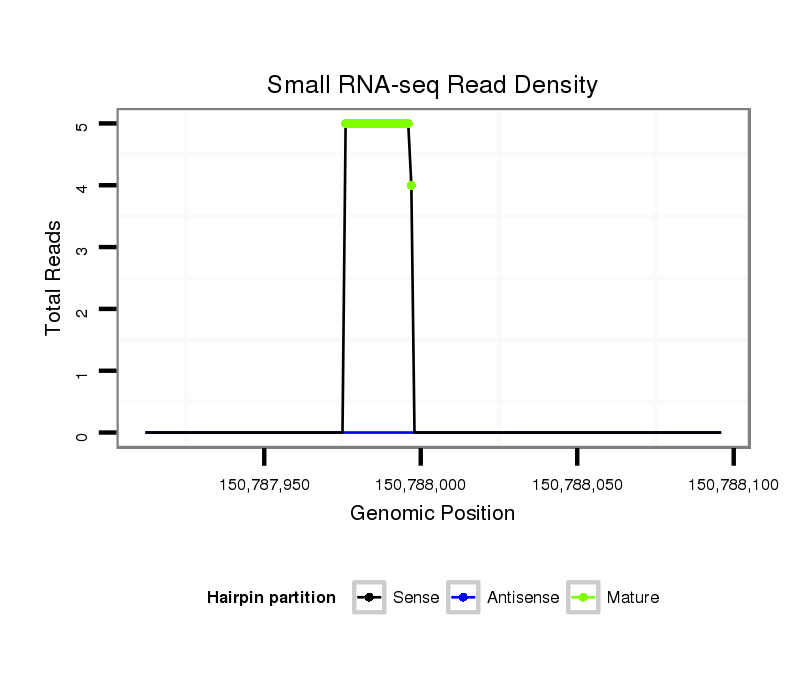
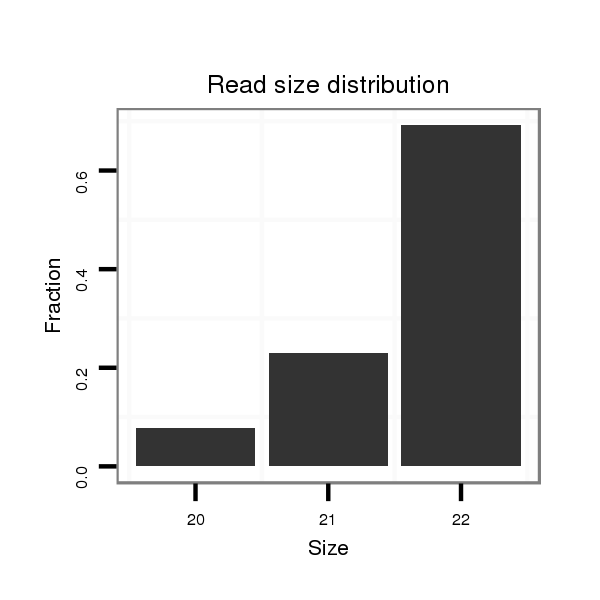
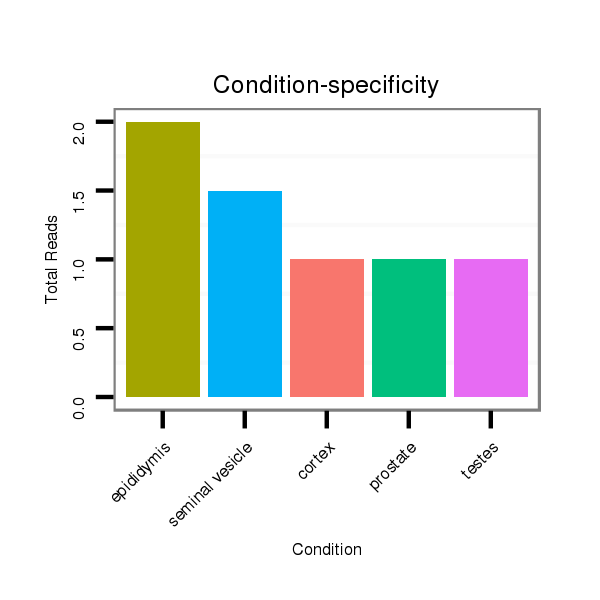
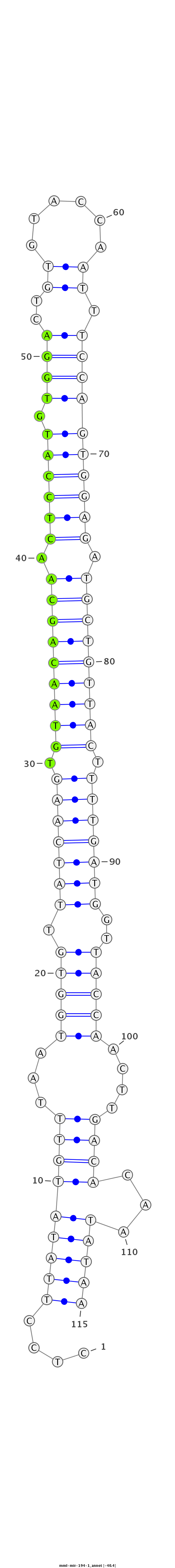
ID:mml-mir-194-1 |
Coordinate:chr1:150787962-150788046 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.6 | -40.5 | -40.4 |
 |
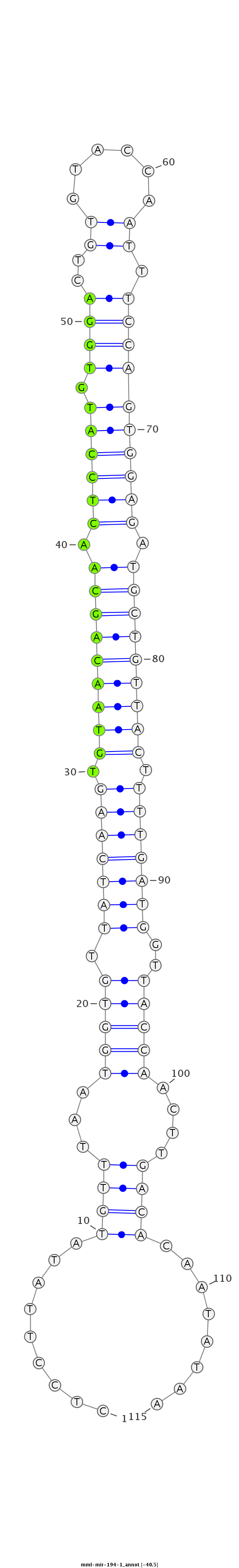 |
 |
| mature | star |
|
GATAATGTACAGATCCCATGATGAGCAAAAGGATTCTCCTTATATGTTTAATGGTGTTATCAAGTGTAACAGCAACTCCATGTGGACTGTGTACCAATTTCCAGTGGAGATGCTGTTACTTTTGATGGTTACCAACTTGACACAATATAAAGGTATAATAAAGAAAGACAGAAAATTGTGGTCAC
***********************************....(((((((((((((((((.(((((((.(((((((((.((((((.((((..((......)).)))))))))).))))))))).)))))))..)))))..)))).)).))))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116845 epididymis |
SRR116844 epididymis |
SRR116840 epididymis |
SRR116839 testes |
SRR116842 seminal vesicle |
SRR116843 cortex |
SRR116841 prostate |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................TGTAACAGCAACTCCATGTGGAATC................................................................................................ | 25 | 2 | 2 | 39.00 | 78 | 53 | 25 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGGATC................................................................................................. | 24 | 2 | 2 | 13.50 | 27 | 21 | 6 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGGA................................................................................................... | 22 | 0 | 2 | 4.50 | 9 | 0 | 0 | 2 | 1 | 2 | 2 | 2 |
| ................................................................TGTAACAGCAACTCCATGTGGAAT................................................................................................. | 24 | 1 | 2 | 3.50 | 7 | 3 | 1 | 0 | 2 | 0 | 1 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGGACTC................................................................................................ | 25 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGGACATC............................................................................................... | 26 | 3 | 2 | 1.50 | 3 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGG.................................................................................................... | 21 | 0 | 2 | 1.50 | 3 | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGGGATC................................................................................................ | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGGAT.................................................................................................. | 23 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................GTAACAGCAACTCCATGTGGAATC................................................................................................ | 24 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGTAACAGCAACTCCATGTGGAATC................................................................................................ | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGGGGAATC................................................................................................ | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATCTGGAATC................................................................................................ | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTGGACG................................................................................................. | 24 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCACCTCCATGTGGATC................................................................................................. | 24 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCAACTCCATGTG..................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TTAACAGCAACTCCATGTGGAATC................................................................................................ | 24 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TGTAACAGCCACTCCATGTGGAATC................................................................................................ | 25 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CTATTACATGTCTAGGGTACTACTCGTTTTCCTAAGAGGAATATACAAATTACCACAATAGTTCACATTGTCGTTGAGGTACACCTGACACATGGTTAAAGGTCACCTCTACGACAATGAAAACTACCAATGGTTGAACTGTGTTATATTTCCATATTATTTCTTTCTGTCTTTTAACACCAGTG
***********************************....(((((((((((((((((.(((((((.(((((((((.((((((.((((..((......)).)))))))))).))))))))).)))))))..)))))..)))).)).))))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:05 PM