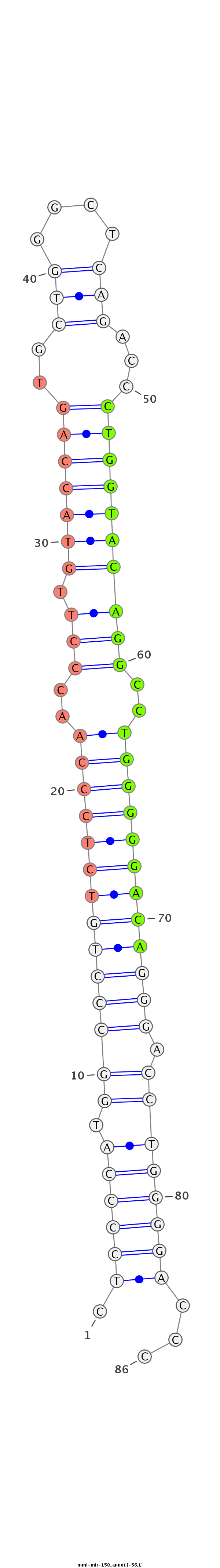
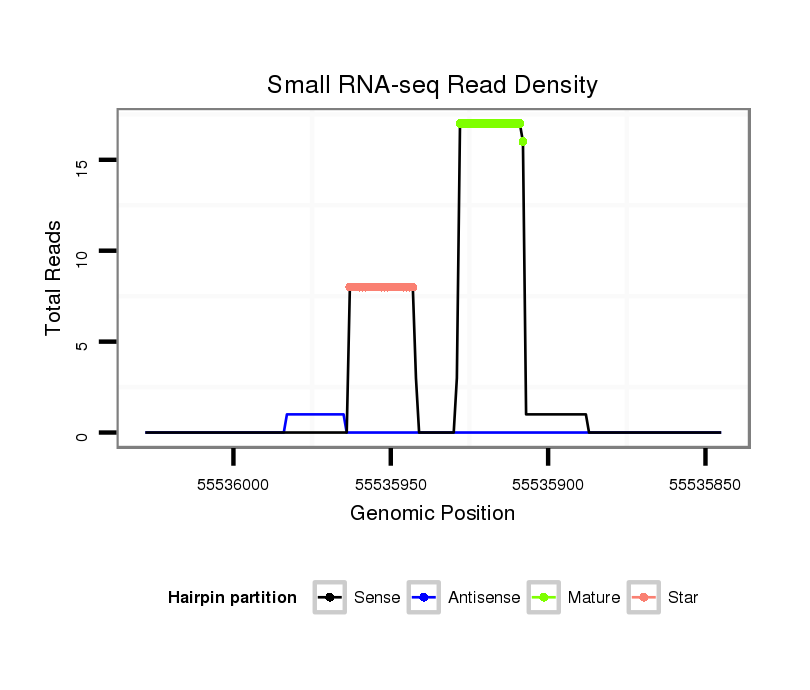
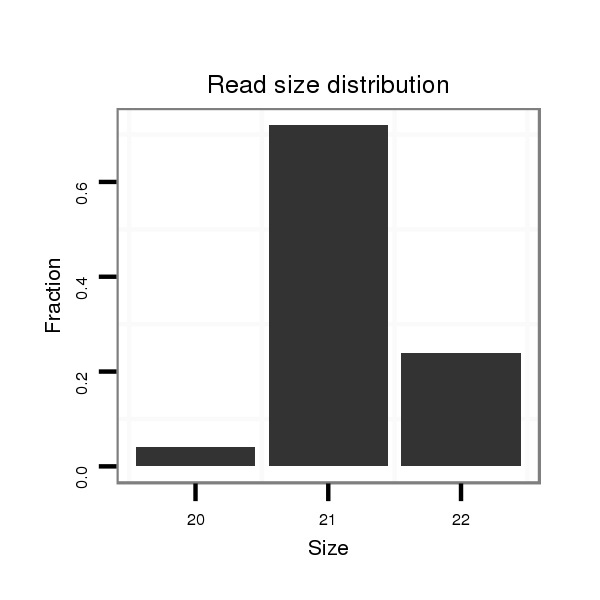
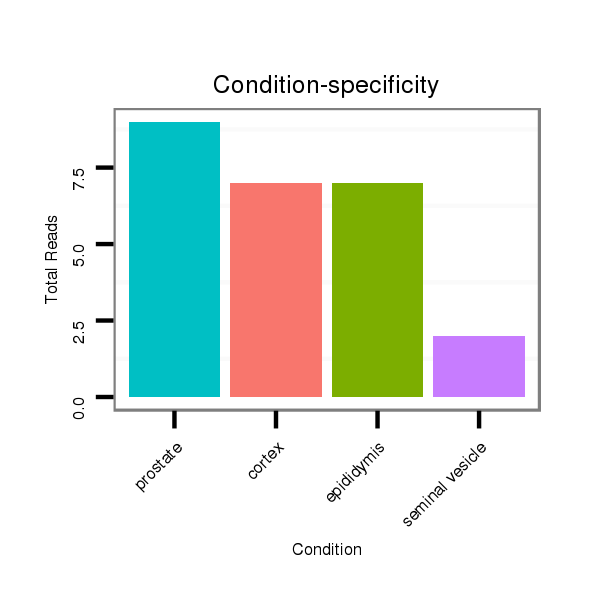
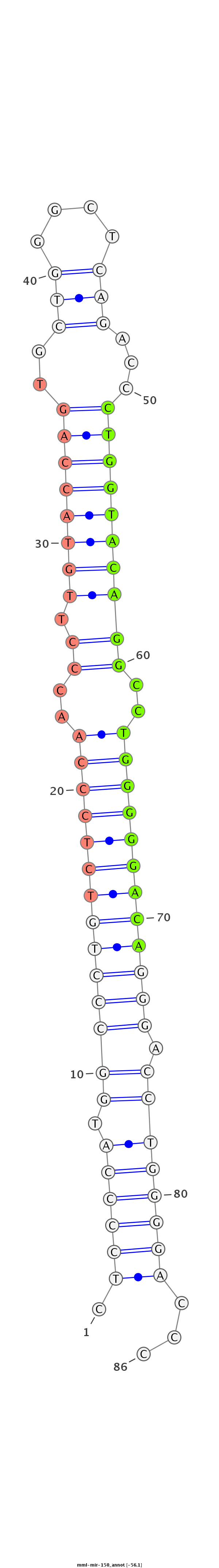
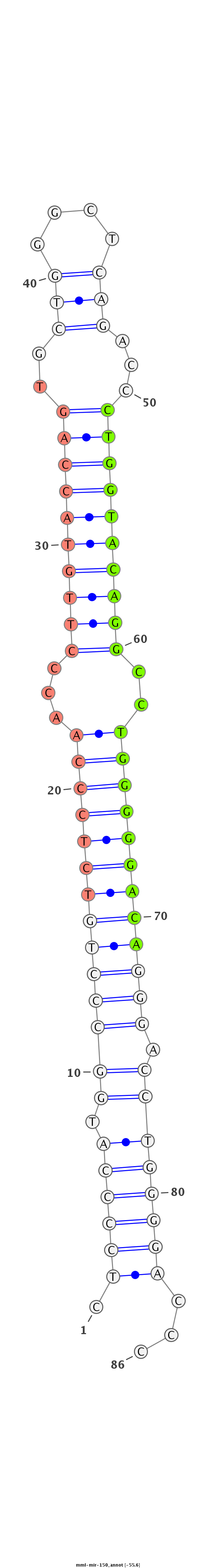
ID:mml-mir-150 |
Coordinate:chr19:55535895-55535978 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -56.1 | -55.6 |
 |
 |
|
GCCCACGGGGAGACGGCGTCCCCGAGACAGCAGCGGCAGCGGCGGCTCCTCTCCCCATGGCCCTGTCTCCCAACCCTTGTACCAGTGCTGGGCTCAGACCCTGGTACAGGCCTGGGGGACAGGGACCTGGGGACCCCGGCACCGGCAGGCCCGAAAGAGGTCAGCAAGCATTCAGACCTCCCTT
**************************************************.((((((.((((((((((((((..(((.(((((((..(((....)))...))))))))))..)))))))))))).))))))))...************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116844 epididymis |
SRR116845 epididymis |
SRR116843 cortex |
SRR116841 prostate |
SRR116840 epididymis |
SRR116842 seminal vesicle |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................TCTCCCAACCCTTGTACCAGTGATC.............................................................................................. | 25 | 2 | 1 | 168.00 | 168 | 128 | 40 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTATC............................................................................................... | 24 | 3 | 2 | 19.50 | 39 | 33 | 6 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACA............................................................... | 21 | 0 | 1 | 13.00 | 13 | 0 | 0 | 3 | 5 | 3 | 2 |
| .................................................................TCTCCCAACCCTTGTACCAGT.................................................................................................. | 21 | 0 | 1 | 5.00 | 5 | 0 | 0 | 1 | 2 | 2 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTGCATC............................................................................................. | 26 | 3 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTGAC............................................................................................... | 24 | 2 | 1 | 4.00 | 4 | 1 | 3 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTAATC.............................................................................................. | 25 | 3 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACAA.............................................................. | 22 | 1 | 1 | 3.00 | 3 | 0 | 0 | 2 | 0 | 0 | 1 |
| ...................................................................................................CCTGGTACAGGCCTGGGGGACA............................................................... | 22 | 0 | 1 | 3.00 | 3 | 0 | 0 | 2 | 0 | 1 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTG................................................................................................. | 22 | 0 | 1 | 3.00 | 3 | 0 | 0 | 0 | 2 | 1 | 0 |
| .........................................................................................................................GGGACCTGGGGACTCCGGCA........................................... | 20 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 1 |
| .................................................................TCTCCCAACCCTTGTACCAGTGTC............................................................................................... | 24 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGATC................................................................................................ | 23 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTCCCAGTGATC.............................................................................................. | 25 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACAT.............................................................. | 22 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| .................................................................TCTCCCAACCCTTTTACCAGTGATC.............................................................................................. | 25 | 3 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCGTTGTACCAGTGATC.............................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTGCTC.............................................................................................. | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTGATA.............................................................................................. | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACAGTT............................................................ | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTTC................................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GGGACCTGGGGACCCCGGCA........................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................CCTGGTACAGGCCTGGGGGACAT.............................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTCATC.............................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGAC................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................TCTCCCACCCCTTGTACCAGTGATC.............................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACATT............................................................. | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTGTTC.............................................................................................. | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACATC............................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTA................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAATC................................................................................................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCATTGATC.............................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCATTGTACCAGTGATC.............................................................................................. | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................CCTGGTACAGGCCTGGGGGACATT............................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................TGTCTCCCAACCCTTGTACCAGTGATC.............................................................................................. | 27 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GGGACCTGGGAACCCCGGCAA.......................................... | 21 | 2 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACAGA............................................................. | 23 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGGGATC.............................................................................................. | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCCTTGTACCAGTTATC.............................................................................................. | 25 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGGTACAGGCCTGGGGGACATA............................................................. | 23 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................TCCCAACCCTTGTACCAGTGCTATC............................................................................................ | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................CCTGGTACAGGCCTGGGGGACAAA............................................................. | 24 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................GGGACCTGGGGACCCCGGCAA.......................................... | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................TGTCTCCCAACCCTTGTACCAGTATC............................................................................................... | 26 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................TCTCCCAACCTTTGTACCAGTGATC.............................................................................................. | 25 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CGGGTGCCCCTCTGCCGCAGGGGCTCTGTCGTCGCCGTCGCCGCCGAGGAGAGGGGTACCGGGACAGAGGGTTGGGAACATGGTCACGACCCGAGTCTGGGACCATGTCCGGACCCCCTGTCCCTGGACCCCTGGGGCCGTGGCCGTCCGGGCTTTCTCCAGTCGTTCGTAAGTCTGGAGGGAA
************************************************.((((((.((((((((((((((..(((.(((((((..(((....)))...))))))))))..)))))))))))).))))))))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116842 seminal vesicle |
SRR116845 epididymis |
|---|---|---|---|---|---|---|---|
| .............................................GAGGAGAGGGGTACCGGGA........................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................................................CTAGGACAGAGGGTTGGGAACATGGT.................................................................................................... | 26 | 2 | 1 | 1.00 | 1 | 0 | 1 |
Generated: 09/01/2015 at 12:15 PM