
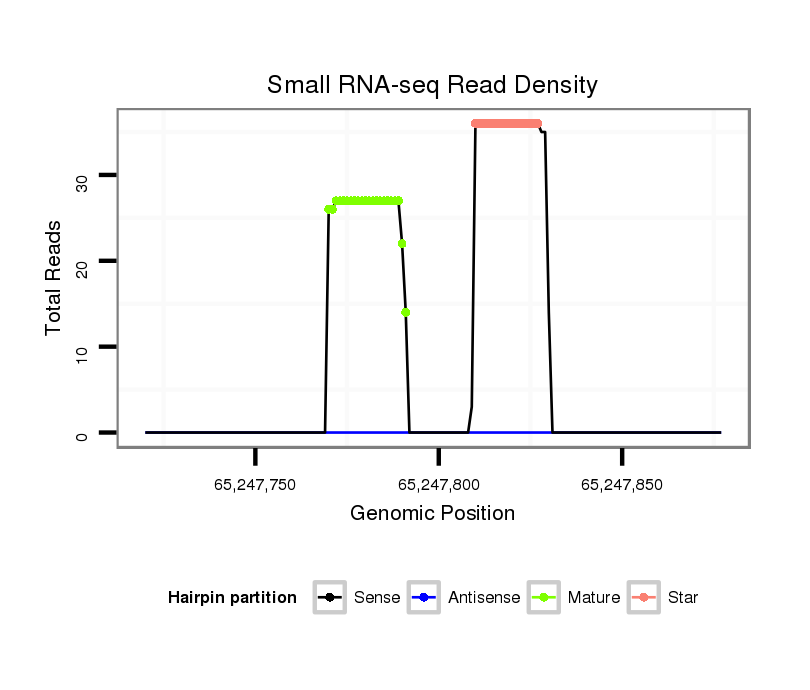
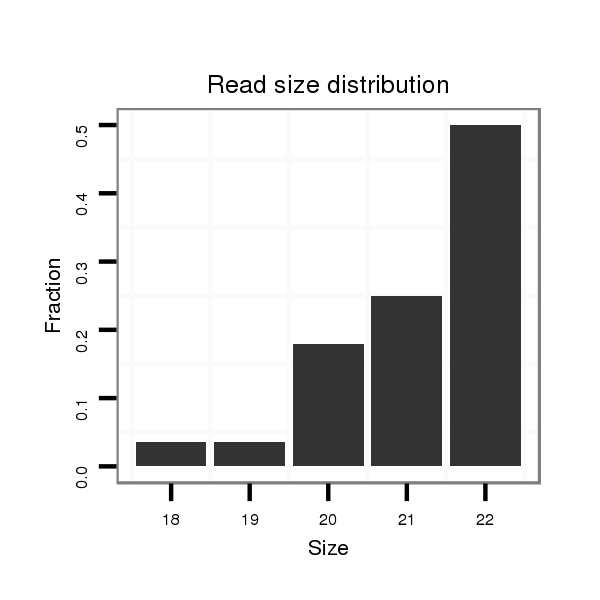
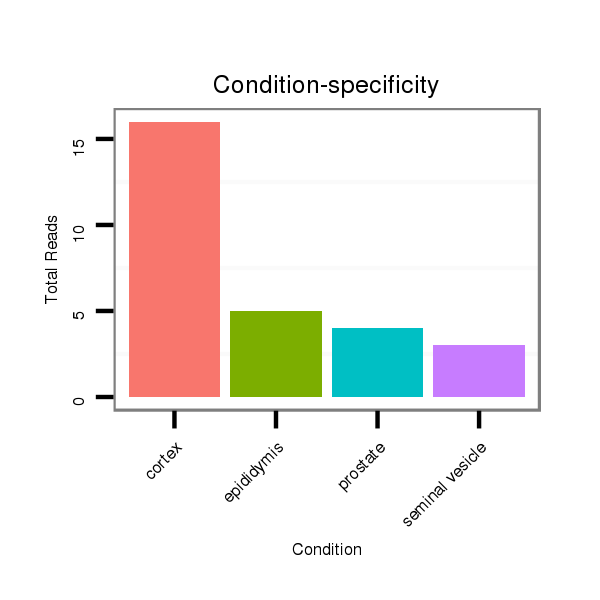
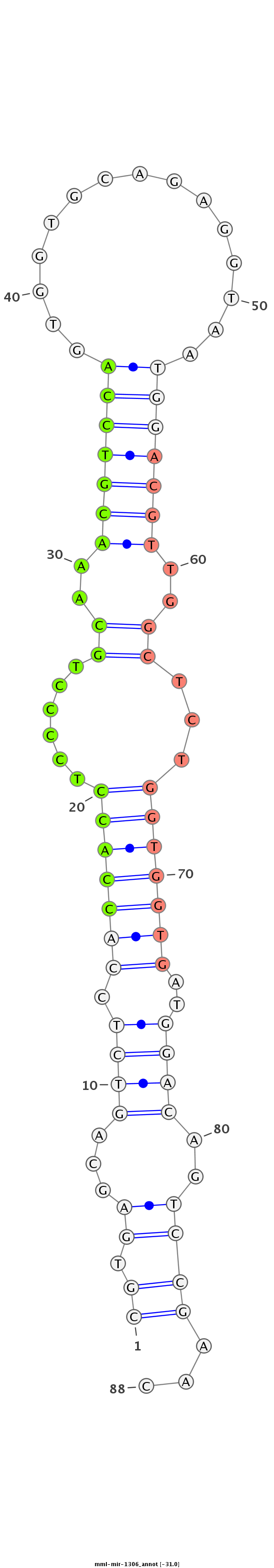
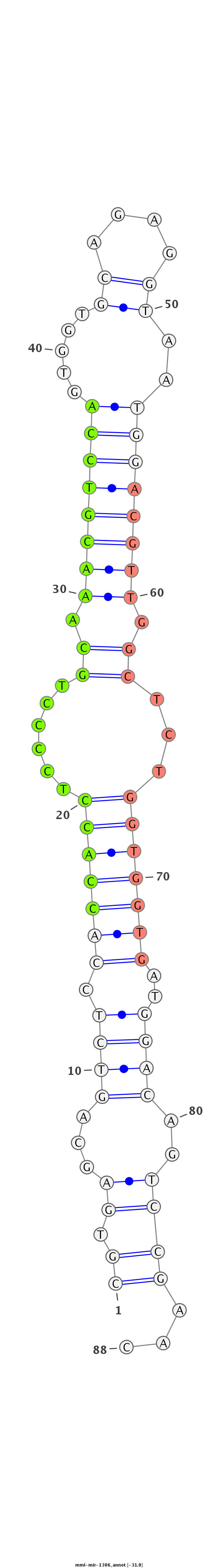
ID:mml-mir-1306 |
Coordinate:chr10:65247770-65247827 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -31.1 | -31.0 | -31.0 |
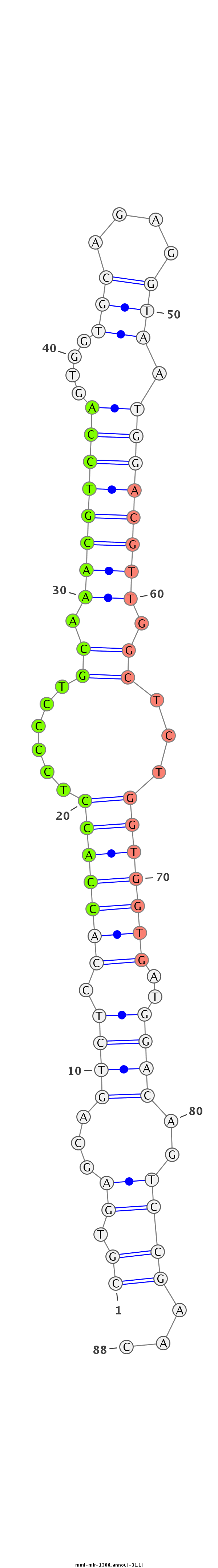 |
 |
 |
| mature | star |
|
TGGAGAGCCGAGCTCGCCCCTTCCAAGCGCTGCCCCGTGAGCAGTCTCCACCACCTCCCCTGCAAACGTCCAGTGGTGCAGAGGTAATGGACGTTGGCTCTGGTGGTGATGGACAGTCCGAACTCCCTGCCGAGGACCCCTTCAACTTCTACGGAGCT
***********************************((.((...((((.(((((((......((.((((((((...............)))))))).))...)))))))..))))..))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116842 seminal vesicle |
SRR116841 prostate |
SRR116840 epididymis |
SRR116845 epididymis |
SRR116839 testes |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................ACGTTGGCTCTGGTGGTGATGT.............................................. | 22 | 1 | 1 | 59.00 | 59 | 27 | 16 | 7 | 6 | 0 | 3 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGAT................................................ | 20 | 0 | 1 | 18.00 | 18 | 8 | 3 | 7 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATG............................................... | 21 | 0 | 1 | 14.00 | 14 | 10 | 3 | 1 | 0 | 0 | 0 | 0 |
| ..................................................CCACCTCCCCTGCAAACGTCCA...................................................................................... | 22 | 0 | 1 | 14.00 | 14 | 8 | 2 | 2 | 2 | 0 | 0 | 0 |
| ..................................................CCACCTCCCCTGCAAACGTCC....................................................................................... | 21 | 0 | 1 | 7.00 | 7 | 4 | 0 | 2 | 1 | 0 | 0 | 0 |
| ..................................................CCACCTCCCCTGCAAACGTCCATC.................................................................................... | 24 | 2 | 1 | 6.00 | 6 | 0 | 0 | 0 | 0 | 3 | 0 | 3 |
| ..................................................CCACCTCCCCTGCAAACGTC........................................................................................ | 20 | 0 | 1 | 5.00 | 5 | 2 | 1 | 0 | 2 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATTT.............................................. | 22 | 2 | 1 | 5.00 | 5 | 1 | 3 | 0 | 1 | 0 | 0 | 0 |
| ..................................................CCACCTCCCCTGCAAACGTCCAATC................................................................................... | 25 | 2 | 1 | 4.00 | 4 | 0 | 0 | 0 | 0 | 3 | 0 | 1 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATT............................................... | 21 | 1 | 1 | 4.00 | 4 | 1 | 1 | 0 | 1 | 0 | 1 | 0 |
| .........................................................................................GACGTTGGCTCTGGTGGTGAT................................................ | 21 | 0 | 1 | 3.00 | 3 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATGA.............................................. | 22 | 1 | 1 | 3.00 | 3 | 0 | 2 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATGTA............................................. | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATGTAT............................................ | 24 | 2 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CCACCTCCCCTGCAAACGTCA....................................................................................... | 21 | 1 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATGAA............................................. | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTCATGT.............................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGCTG............................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................AAGACGTTGGCTCTGGTGGTGAT................................................ | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTTAT................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................ACCTCCCCTGCAAACGTCC....................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATAT.............................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTG.................................................. | 18 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATTTA............................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATTA.............................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CCACCTCCCCTGCAAACGTCCAT..................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTTATG............................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGGGATG............................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATTTAT............................................ | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGTTGG.............................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATGAAA............................................ | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATGTTT............................................ | 24 | 3 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATAA.............................................. | 22 | 2 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ACGTTGGCTCTGGTGGTGATTTT............................................. | 23 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................CACCTCCCCTGCAAACGTCCATC.................................................................................... | 23 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................CCCCCTCCCCTGCAAACGTCCATC.................................................................................... | 24 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................CCACCTCCCCTGCAAACGTCATC..................................................................................... | 23 | 3 | 12 | 0.25 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| ..........................................................................................ACGTTGGCTCTGGTGGTTATTT.............................................. | 22 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................CACCTCCCCTGCAAACGTCATC..................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ACCTCTCGGCTCGAGCGGGGAAGGTTCGCGACGGGGCACTCGTCAGAGGTGGTGGAGGGGACGTTTGCAGGTCACCACGTCTCCATTACCTGCAACCGAGACCACCACTACCTGTCAGGCTTGAGGGACGGCTCCTGGGGAAGTTGAAGATGCCTCGA
***********************************((.((...((((.(((((((......((.((((((((...............)))))))).))...)))))))..))))..))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116844 epididymis |
|---|---|---|---|---|---|---|
| ...........................................................................................................CTACCTGTTAGGCTTGA.................................. | 17 | 1 | 2 | 1.50 | 3 | 3 |
| ...........................................................................................................CTACCTGTTAGGCTTG................................... | 16 | 1 | 6 | 0.17 | 1 | 1 |
Generated: 09/01/2015 at 12:06 PM