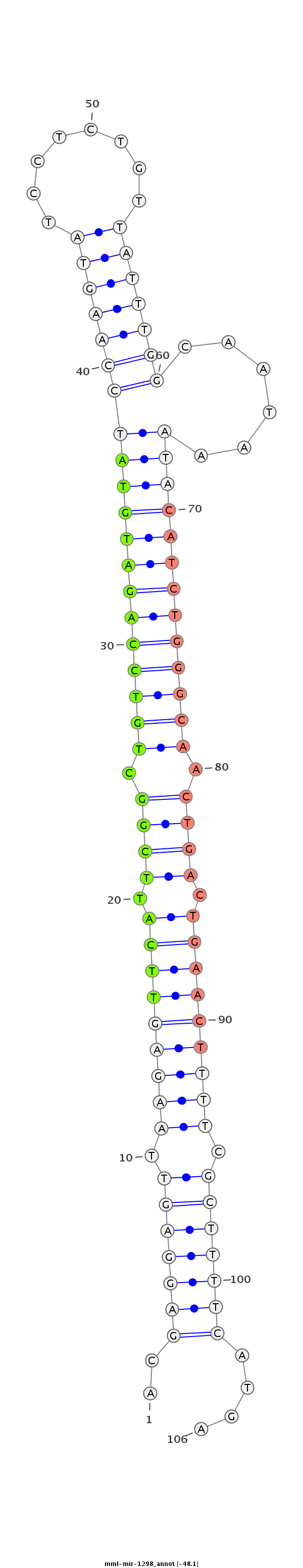
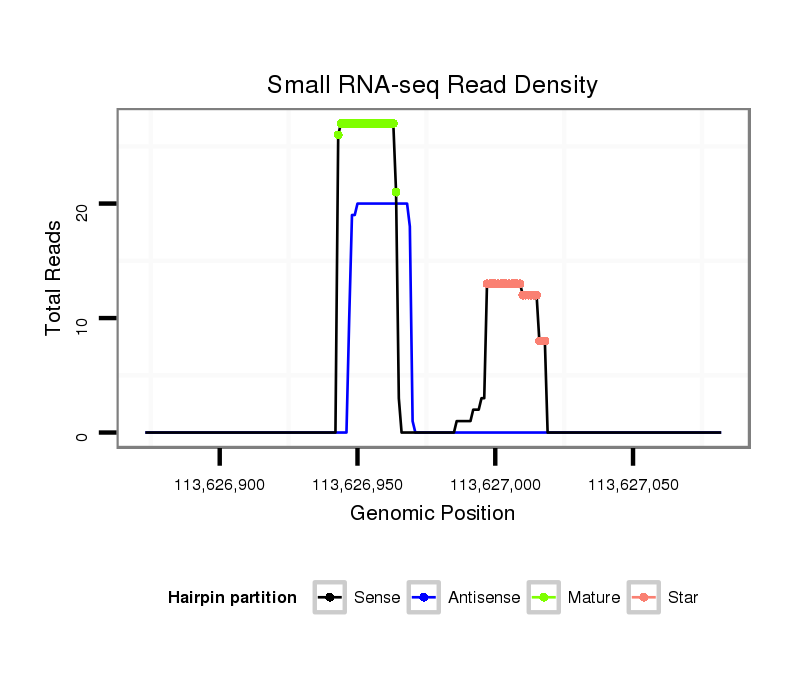
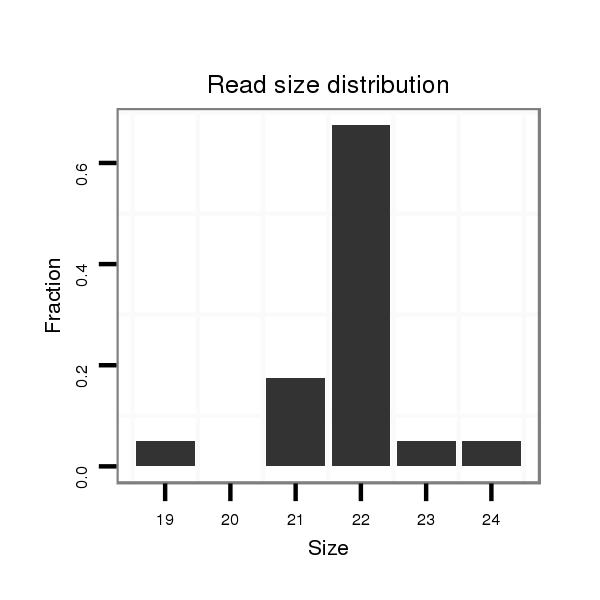
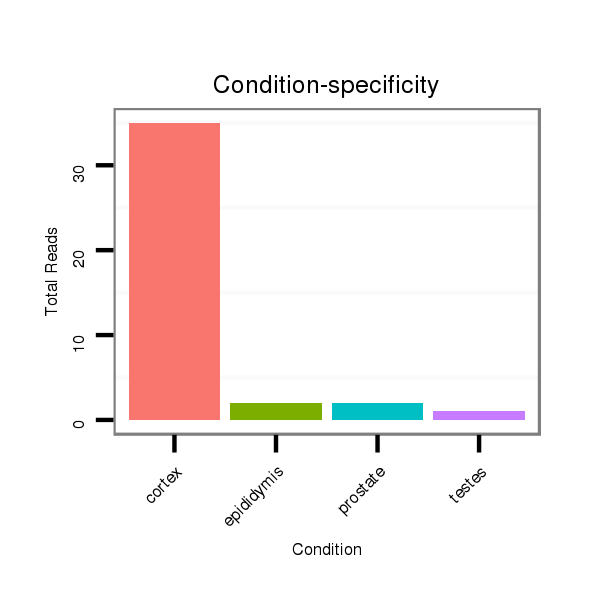
ID:mml-mir-1298 |
Coordinate:chrX:113626923-113627032 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
TCTTTCCCCTTCATATAGGATGTCTTGAAGCTGAGAACGAAGAGCTGTTGGAGAGACGAGGAGTTAAGAGTTCATTCGGCTGTCCAGATGTATCCAAGTATCCTCTGTTATTTGGCAATAAATACATCTGGGCAACTGACTGAACTTTTCGCTTTTCATGACTCAGCTGGAACCCTCCACAACCAGAAGAGCCTGTTATAAAGTGAAAAG
*******************************************************..(((((((.(((((((((.((((.((((((((((((((((((((........)))))))......))))))))))))).)))).))))))))).)))))))....************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116843 cortex |
SRR116840 epididymis |
SRR116841 prostate |
SRR116839 testes |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TTCATTCGGCTGTCCAGATGTA...................................................................................................................... | 22 | 0 | 1 | 18.00 | 18 | 16 | 1 | 1 | 0 | 0 |
| ............................................................................................................................CATCTGGGCAACTGACTGAACT................................................................ | 22 | 0 | 1 | 8.00 | 8 | 7 | 0 | 1 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATGT....................................................................................................................... | 21 | 0 | 1 | 6.00 | 6 | 5 | 1 | 0 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATTTA...................................................................................................................... | 22 | 1 | 1 | 4.00 | 4 | 2 | 1 | 1 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATTTAA..................................................................................................................... | 23 | 2 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATGTAT..................................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................................................................CATCTGGGCAACTGACTGA................................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATGTAA..................................................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................CATCTGGGCAACTGACTGAAA................................................................. | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................TCATTCGGCTGTCCAGATGTAA..................................................................................................................... | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TACATCTGGGCAACTGACTGAACAA............................................................... | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCATATGAA...................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TACATCTGGGCAACTGACTGA................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AAATACATCTGGGCAACTGACTGA................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CATCTGGGCAACTGACTGAACTA............................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCCGATGTA...................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................GGCAATAAATACATCTGGGCAACT......................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................TCATTCGGCTGTCCAGATGTAT..................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CATCTGGGCAACTGACTGAACAA............................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATT........................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATTTATC.................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................TTCATTCGGCTGTCCAGATTTAT..................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................ATACATCTGGGCAACTGACTTA................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................TAAGAGTTCGTTCGACTATC.............................................................................................................................. | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
|
AGAAAGGGGAAGTATATCCTACAGAACTTCGACTCTTGCTTCTCGACAACCTCTCTGCTCCTCAATTCTCAAGTAAGCCGACAGGTCTACATAGGTTCATAGGAGACAATAAACCGTTATTTATGTAGACCCGTTGACTGACTTGAAAAGCGAAAAGTACTGAGTCGACCTTGGGAGGTGTTGGTCTTCTCGGACAATATTTCACTTTTC
*************************************************..(((((((.(((((((((.((((.((((((((((((((((((((........)))))))......))))))))))))).)))).))))))))).)))))))....******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116842 seminal vesicle |
SRR116844 epididymis |
|---|---|---|---|---|---|---|---|---|
| ...........................................................................AGCCGACAGGTCTACATAGGTT................................................................................................................. | 22 | 0 | 1 | 9.00 | 9 | 8 | 1 | 0 |
| ..........................................................................AAGCCGACAGGTCTACATAGGTT................................................................................................................. | 23 | 0 | 1 | 7.00 | 7 | 6 | 1 | 0 |
| ..........................................................................AAGCCGACAGGTCTACATAGGT.................................................................................................................. | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ...........................................................................TGCCGACAGGTCTACATAGGTT................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................GACGACAGGTCTACATAGGTT................................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .............................................................................CCGACAGGTCTACATAGGTT................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................AAGCCGACAGGTCTACATAGGTTC................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................AGCCGACAGTTCTACATAGGTT................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................TAGCCGACAGGTCTACATAGGTT................................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................................................AAAGCCGACAGGTCTACATAGGT.................................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................CAAGTCTCAAGTAGGCCG.................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..............................................................CAAGTCTCAAGTAAGC.................................................................................................................................... | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 |
Generated: 09/01/2015 at 12:34 PM