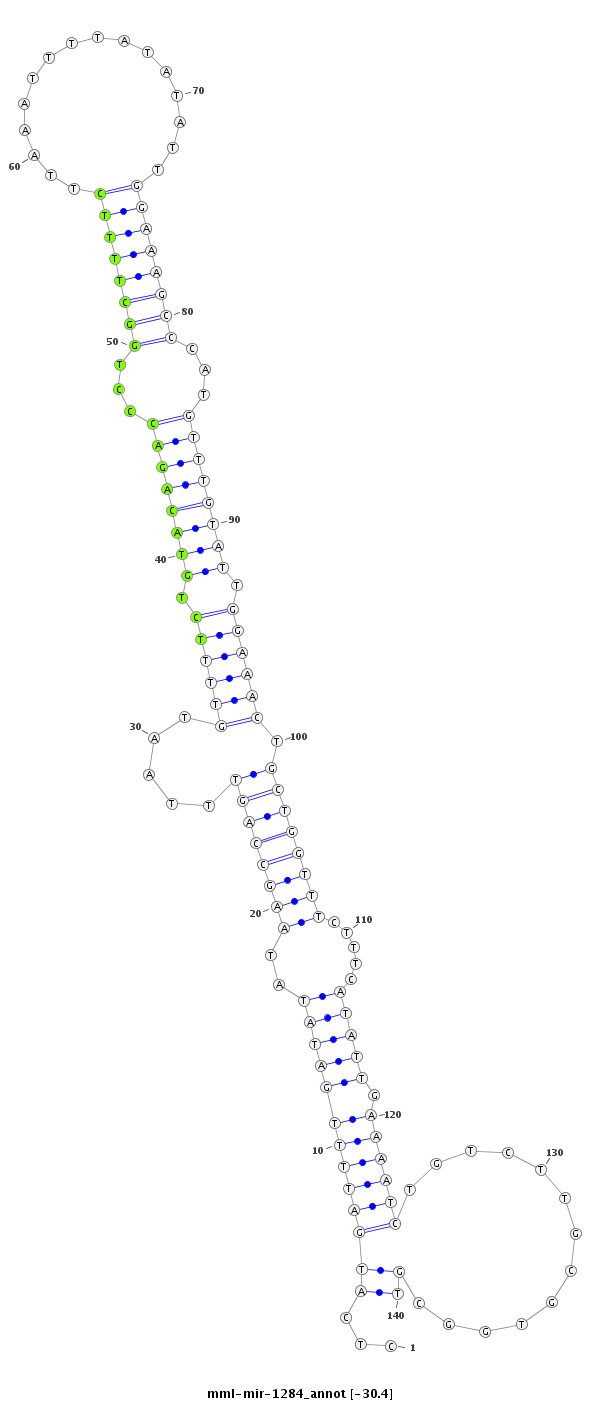
ID:mml-mir-1284 |
Coordinate:chr2:66131428-66131537 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
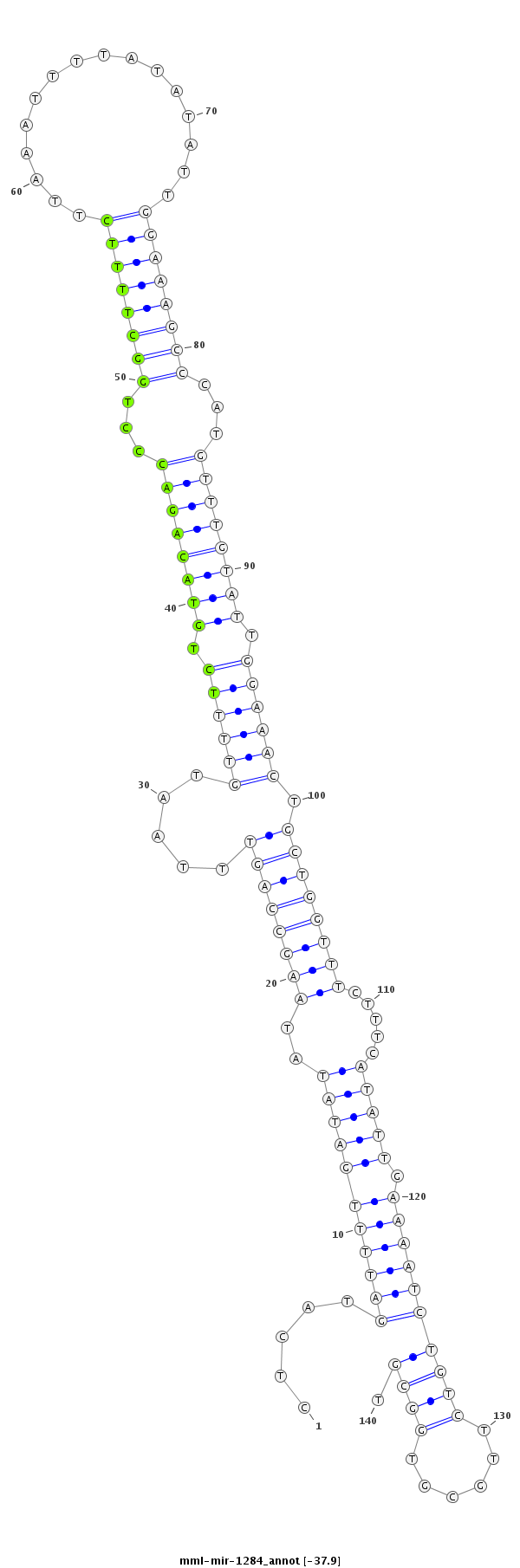
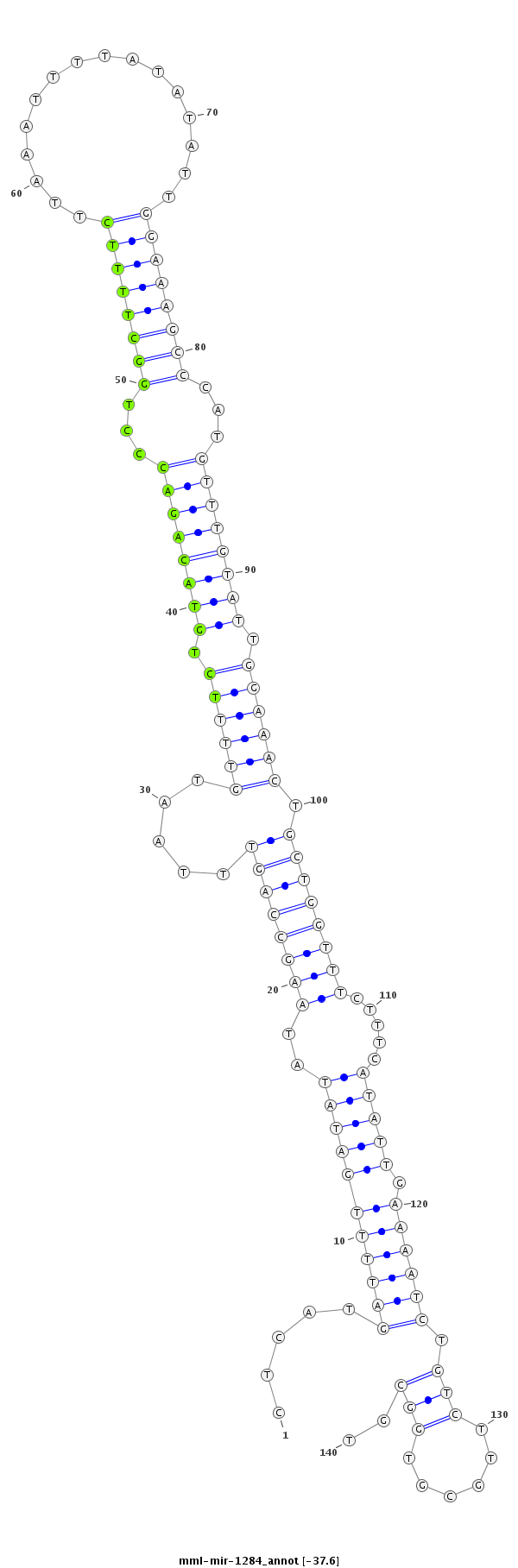
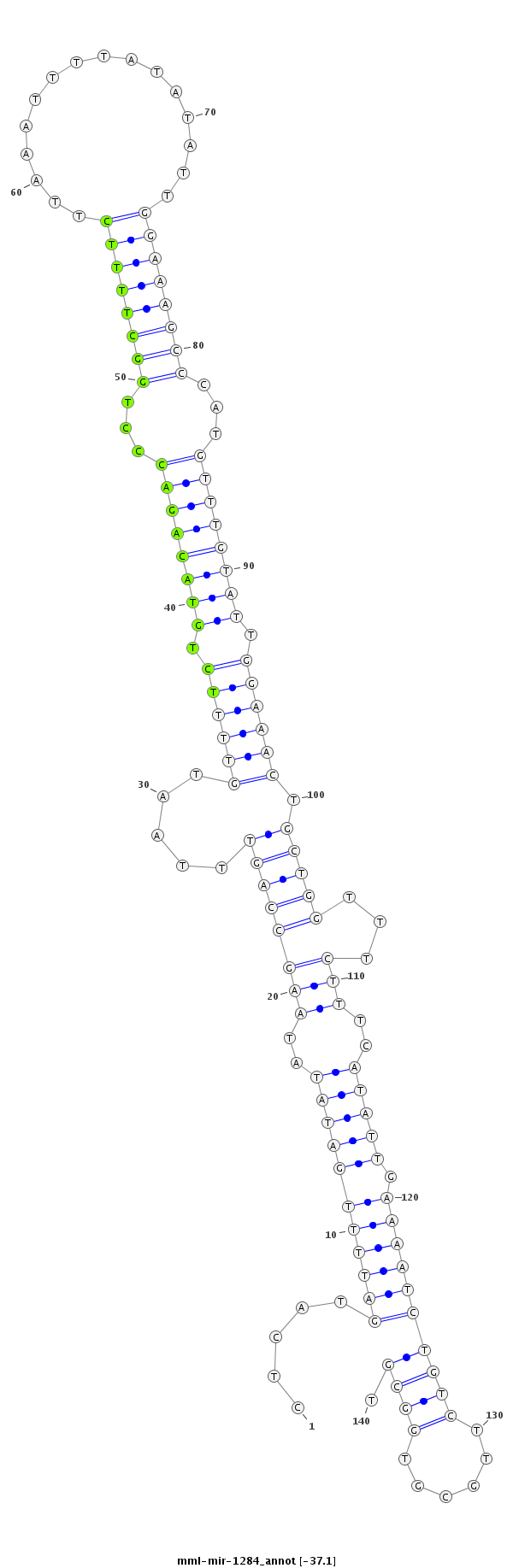
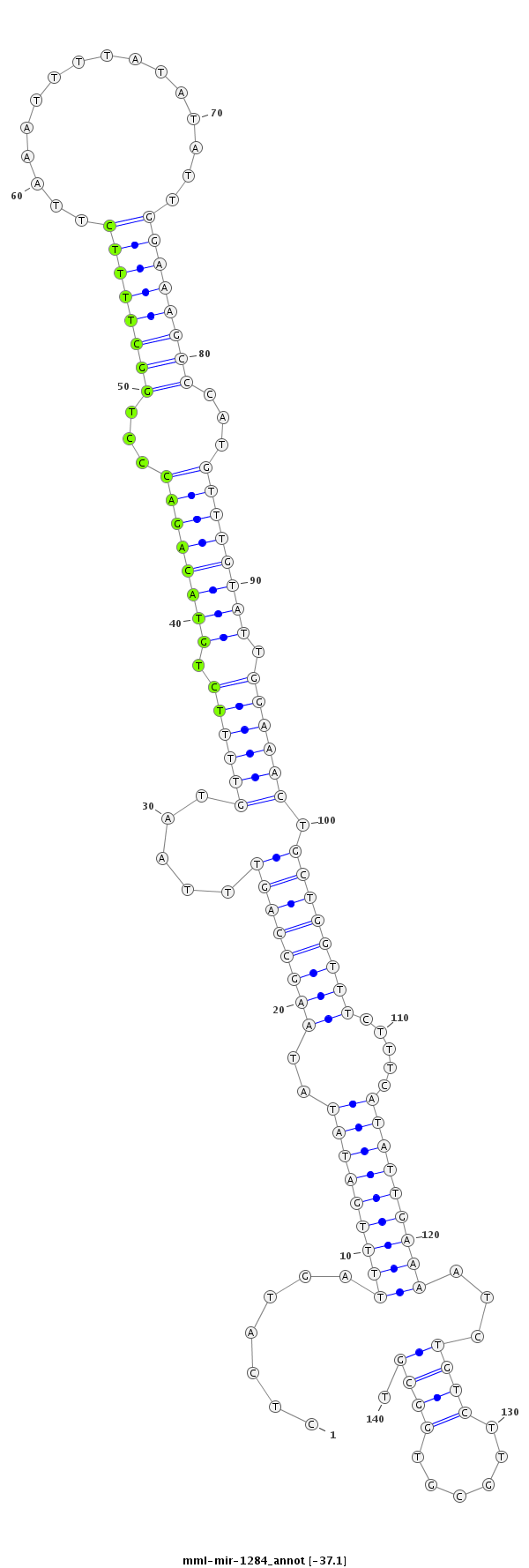
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -37.9 | -37.6 | -37.1 | -37.1 |
 |
 |
 |
 |
|
TGTAAATTATTTTAATGCTGCATTTCTGCCCAAACCTCATGATTTTGATATATAAGCCAGTTTAATGTTTTCTGTACAGACCCTGGCTTTTCTTAAATTTTATATATTGGAAAGCCCATGTTTGTATTGGAAACTGCTGGTTTCTTTCATATTGAAAATCTGTCTTGCGTGGCGTGATGCCATGTGGCAAAGGAGTAGTTTGACCAGAGT
***********************************...(((((((((((((..((((((((.....((((((.((((((((...((((((((................))))))))...)))))))).)))))).)))))))).....))))).)))))).............))*********************************** |
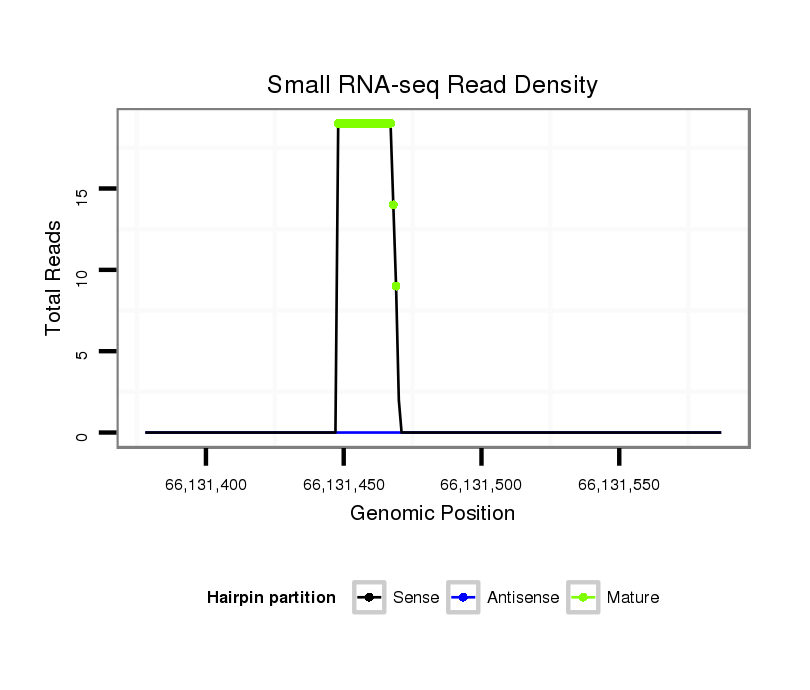
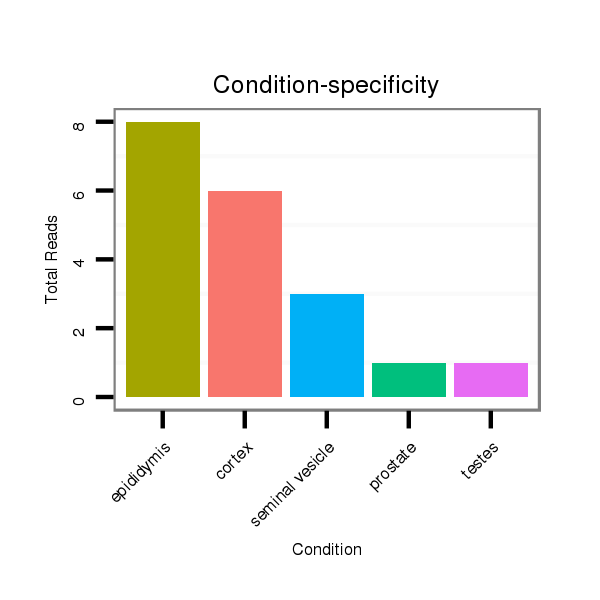
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116843 cortex |
SRR116842 seminal vesicle |
SRR116839 testes |
SRR116841 prostate |
|---|---|---|---|---|---|---|---|---|---|---|
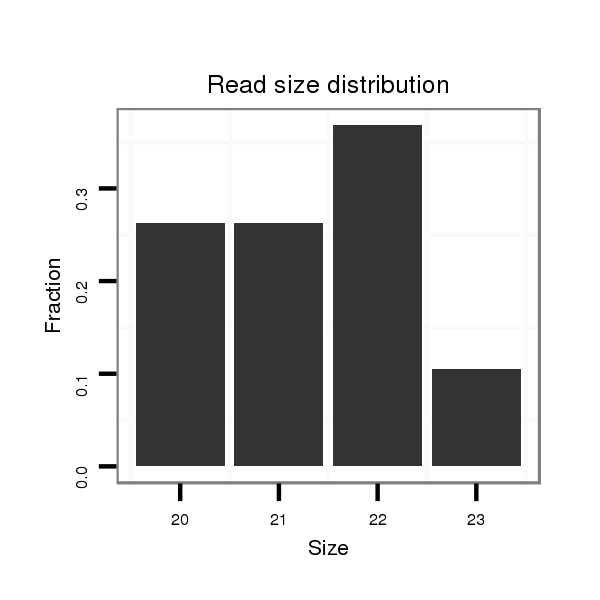
| ......................................................................TCTGTACAGACCCTGGCTTTTC...................................................................................................................... | 22 | 0 | 1 | 7.00 | 7 | 2 | 4 | 0 | 1 | 0 |
| ......................................................................TCTGTACAGACCCTGGCTTTT....................................................................................................................... | 21 | 0 | 1 | 5.00 | 5 | 4 | 0 | 1 | 0 | 0 |
| ......................................................................TCTGTACAGACCCTGGCTTT........................................................................................................................ | 20 | 0 | 1 | 5.00 | 5 | 2 | 2 | 1 | 0 | 0 |
| ......................................................................TCTGTACAGACCCTGGCTTTTCT..................................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 1 |
| ......................................................................TCTGTACAGACCCTGGCTTTTCA..................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................TCTGTACAGACCCTGGCTTTA....................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................TCTGTACAGCCCCTGGCTTT........................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................TCTGTACAGACCCTGGCTAA........................................................................................................................ | 20 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
|
ACATTTAATAAAATTACGACGTAAAGACGGGTTTGGAGTACTAAAACTATATATTCGGTCAAATTACAAAAGACATGTCTGGGACCGAAAAGAATTTAAAATATATAACCTTTCGGGTACAAACATAACCTTTGACGACCAAAGAAAGTATAACTTTTAGACAGAACGCACCGCACTACGGTACACCGTTTCCTCATCAAACTGGTCTCA
***********************************...(((((((((((((..((((((((.....((((((.((((((((...((((((((................))))))))...)))))))).)))))).)))))))).....))))).)))))).............))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:19 PM