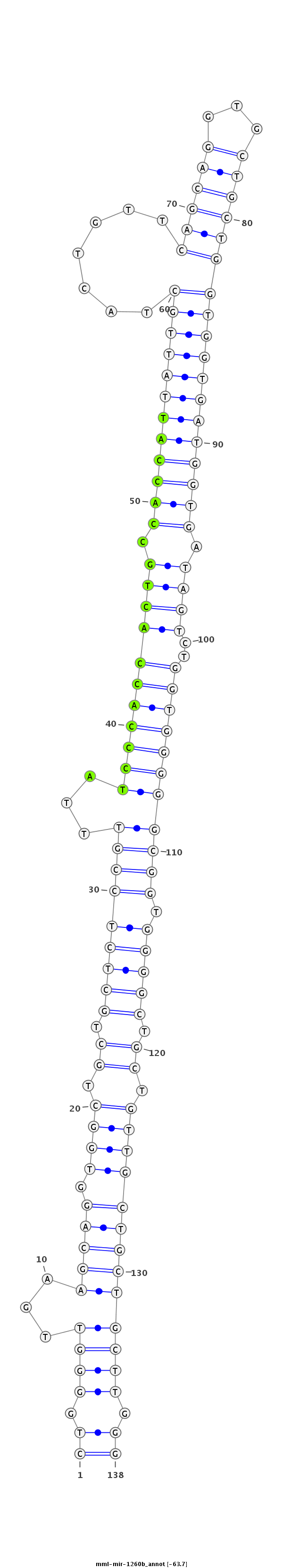

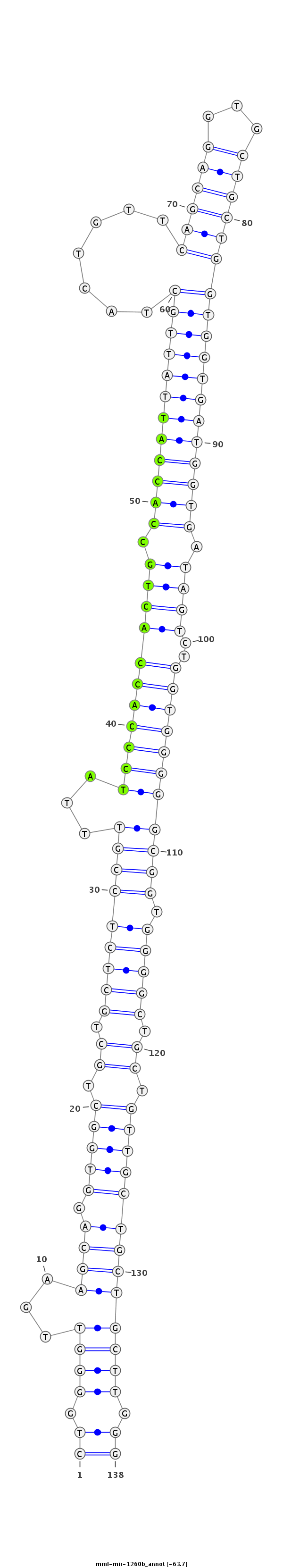
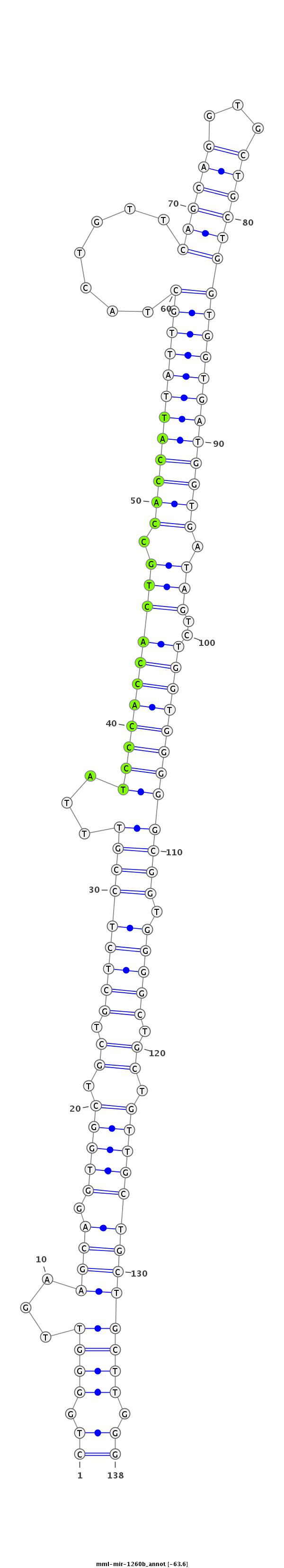
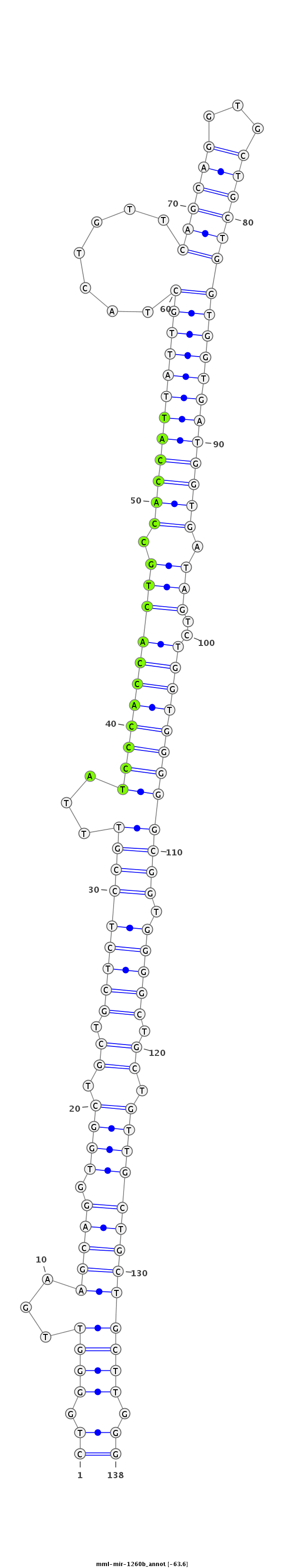
ID:mml-mir-1260b |
Coordinate:chr14:97339654-97339761 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -63.7 | -63.6 | -63.6 |
 |
 |
 |
|
GCTTACCGCAATCAGGGCTGAGTTCCTCTGGTCCCCTGGGGTTGAAGCAGGTGGCTGCTGCTCTCCGTTTATCCCACCACTGCCACCATTATTGCTACTGTTCAGCAGGTGCTGCTGGTGGTGATGGTGATAGTCTGGTGGGGGCGGTGGGGCTGCTGTTGCTGCTGCTTGGGAGGCCGCAGGAGGGGCAGCAGGGGGCGGTGGAGGG
***********************************((.((((...(((((.((((.((.(((((((((...(((((((((((.((((((((((((.......((((((...)))))))))))))))))).))))..))))))))))).))))).)).))))))))))))).))*********************************** |
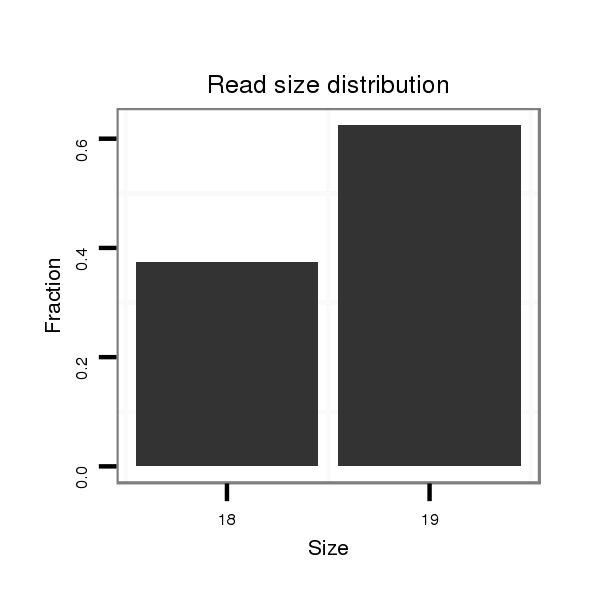
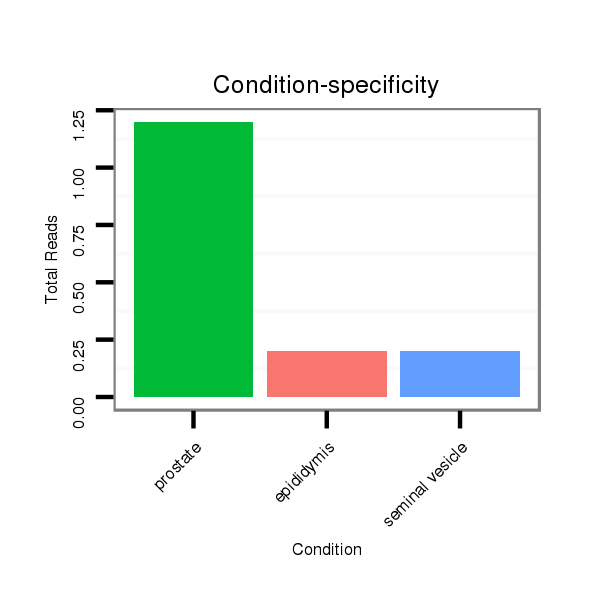
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116842 seminal vesicle |
SRR116845 epididymis |
SRR116841 prostate |
SRR116844 epididymis |
SRR116840 epididymis |
SRR116843 cortex |
SRR116839 testes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................ATCCCACCGCTGCCACCATT...................................................................................................................... | 20 | 1 | 1 | 7.00 | 7 | 3 | 0 | 2 | 0 | 1 | 1 | 0 |
| ......................................................................ATCCCACCGCTGCCACCA........................................................................................................................ | 18 | 1 | 20 | 6.40 | 128 | 60 | 0 | 39 | 0 | 25 | 4 | 0 |
| ......................................................................ATCCCACCGCTGCCACCATTATC................................................................................................................... | 23 | 2 | 2 | 4.00 | 8 | 0 | 5 | 0 | 3 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCAT....................................................................................................................... | 19 | 1 | 2 | 3.00 | 6 | 4 | 0 | 0 | 0 | 0 | 1 | 1 |
| ......................................................................ATCCCACCGCTGCCACCATTAATC.................................................................................................................. | 24 | 3 | 1 | 3.00 | 3 | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCATC...................................................................................................................... | 20 | 2 | 20 | 1.30 | 26 | 0 | 15 | 0 | 11 | 0 | 0 | 0 |
| ......................................................................ATCCCACCACTGCCACCAT....................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................ATCCCACCACTGCCACCATTATC................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCAATATC................................................................................................................... | 23 | 3 | 11 | 0.73 | 8 | 0 | 5 | 0 | 3 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCATAAT.................................................................................................................... | 22 | 2 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ATCCCACCACTGCCACCA........................................................................................................................ | 18 | 0 | 5 | 0.60 | 3 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACC......................................................................................................................... | 17 | 1 | 20 | 0.50 | 10 | 7 | 0 | 3 | 0 | 0 | 0 | 0 |
| .......................................................................TCCCACCGCTGCCACCATTAATTC................................................................................................................. | 24 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCATAATC................................................................................................................... | 23 | 3 | 7 | 0.43 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ......................................................................ATCCCACCACTGCCACCATAATC................................................................................................................... | 23 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCATTAA.................................................................................................................... | 22 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TATCCCACCGCTGCCACCA........................................................................................................................ | 19 | 1 | 7 | 0.29 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................ATCCCACCACTGCCACCAATC..................................................................................................................... | 21 | 2 | 15 | 0.27 | 4 | 0 | 2 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................TCCCACCGCTGCCACCATT...................................................................................................................... | 19 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCATTC..................................................................................................................... | 21 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................ATCCCACCGCTGCCACCATA...................................................................................................................... | 20 | 2 | 18 | 0.11 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................TCCCACCGCTGCCACCAT....................................................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................ATCCGACCGCTGCCACCA........................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CGAATGGCGTTAGTCCCGACTCAAGGAGACCAGGGGACCCCAACTTCGTCCACCGACGACGAGAGGCAAATAGGGTGGTGACGGTGGTAATAACGATGACAAGTCGTCCACGACGACCACCACTACCACTATCAGACCACCCCCGCCACCCCGACGACAACGACGACGAACCCTCCGGCGTCCTCCCCGTCGTCCCCCGCCACCTCCC
***********************************((.((((...(((((.((((.((.(((((((((...(((((((((((.((((((((((((.......((((((...)))))))))))))))))).))))..))))))))))).))))).)).))))))))))))).))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:10 PM