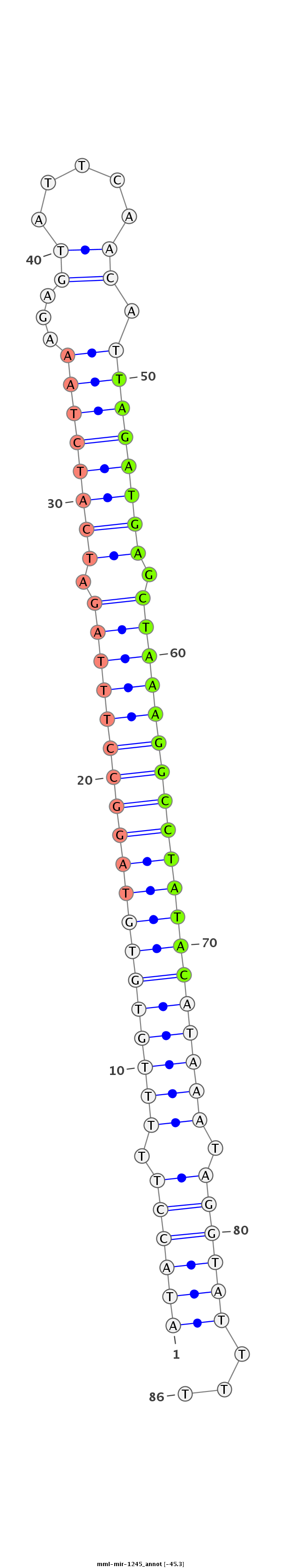
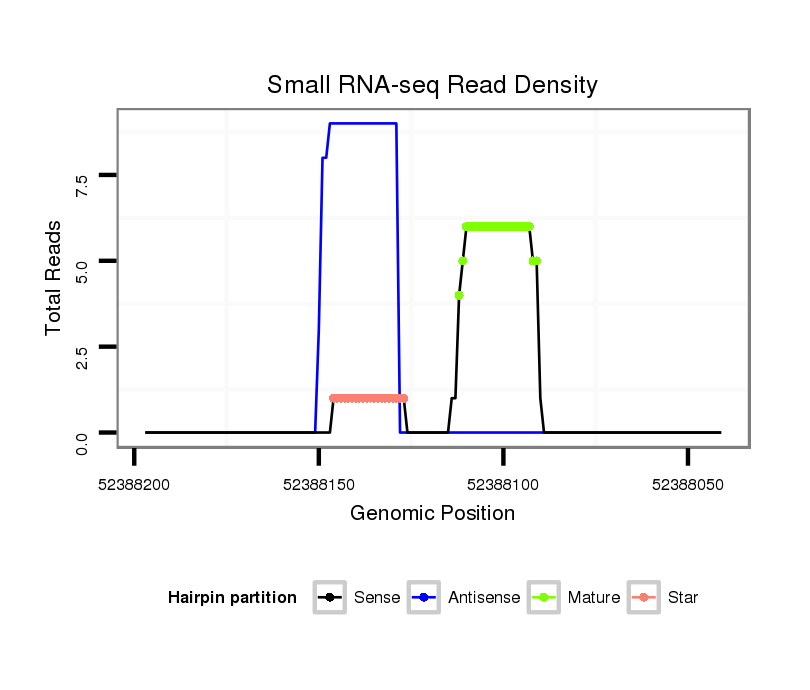
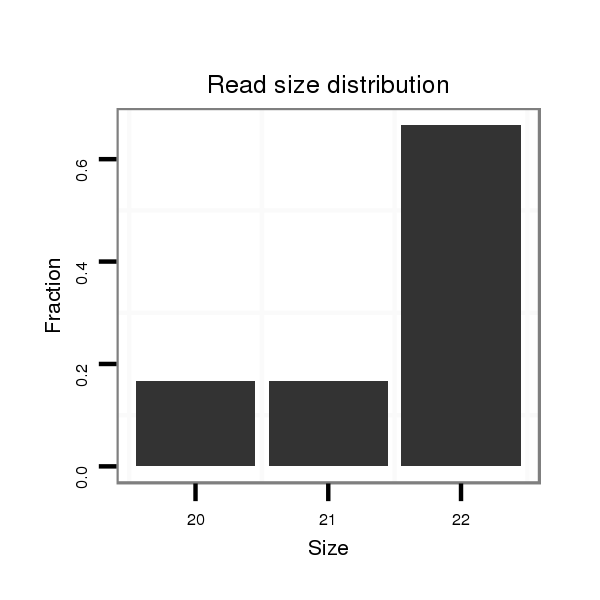
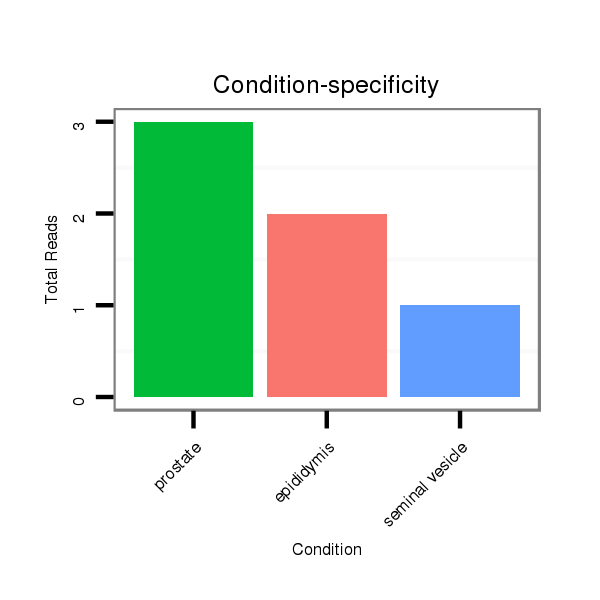
ID:mml-mir-1245 |
Coordinate:chr12:52388091-52388147 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -44.8 | -44.4 | -44.3 |
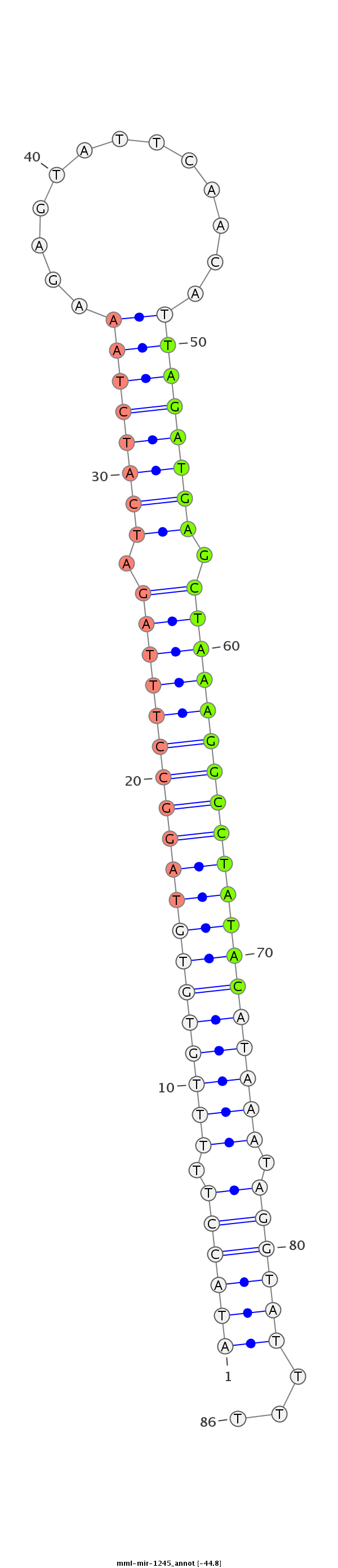 |
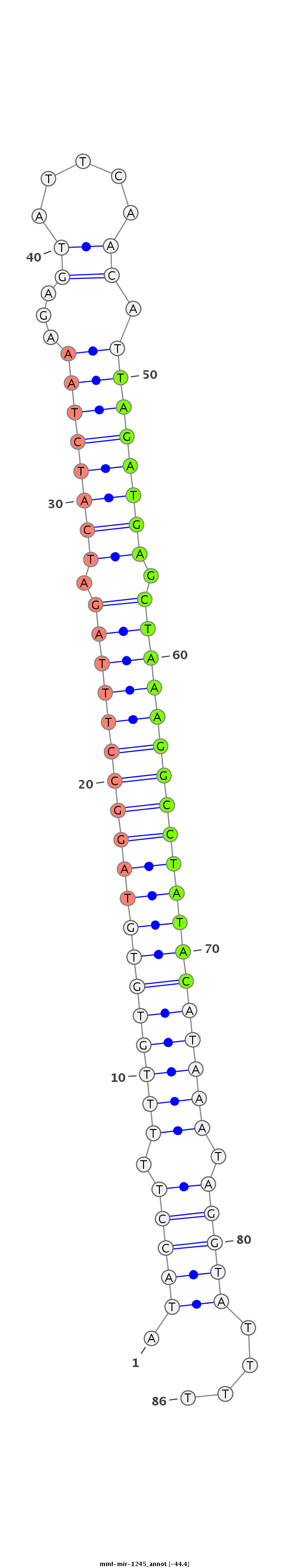 |
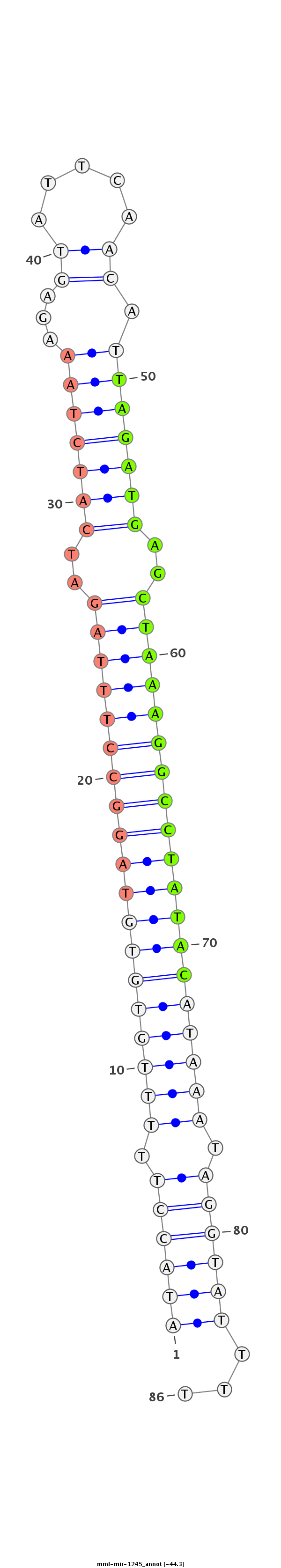 |
|
ATGCTGAATTTTATTTAATTCCAGAATTTAATAAAAATACCTTTTTGTGTGTAGGCCTTTAGATCATCTAAAGAGTATTCAACATTAGATGAGCTAAAGGCCTATACATAAATAGGTATTTTTGAAAATCCAAATTTTTCAAAGCTCTGGTATTACC
************************************((((((.(((((((((((((((((((.((((((((...((.....)).)))))))).))))))))))))))))))).))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116840 epididymis |
SRR116844 epididymis |
SRR116841 prostate |
SRR116842 seminal vesicle |
SRR116845 epididymis |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................TAGATGAGCTAAAGGCCTATAC.................................................. | 22 | 0 | 1 | 3.00 | 3 | 1 | 0 | 2 | 0 | 0 |
| ..............................................ATGTGTAGGCCTTTAGATCAT.......................................................................................... | 21 | 1 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 | 0 |
| ..................................................GTAGGCCTTTAGATCATCTAATC.................................................................................... | 23 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...................................................TAGGCCTTTAGATCATCTAA...................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................TATGTGTAGGCCTTTAGATCATC......................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................AGATGAGCTAAAGGCCTATAC.................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................TAGGCCTTTAGATCATCTAAATC................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GTAGGCCTTTAGATCATCTAAATC................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................GATGAGCTAAAGGCCTATACA................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................ATTAGATGAGCTAAAGGCCTAT.................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................TATGTGTAGGCCTTTAGATCAT.......................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................TGTAGGCCTTTAGATCATCTAAATC................................................................................... | 25 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................TAGATGAGCTAAAGGCCTCTAC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GTAGGCCTTTAGACCATC......................................................................................... | 18 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................TAGGCCTTTAGATCATCA........................................................................................ | 18 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TACGACTTAAAATAAATTAAGGTCTTAAATTATTTTTATGGAAAAACACACATCCGGAAATCTAGTAGATTTCTCATAAGTTGTAATCTACTCGATTTCCGGATATGTATTTATCCATAAAAACTTTTAGGTTTAAAAAGTTTCGAGACCATAATGG
***********************************((((((.(((((((((((((((((((.((((((((...((.....)).)))))))).))))))))))))))))))).))))))...************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR116841 prostate |
SRR116840 epididymis |
SRR116842 seminal vesicle |
|---|---|---|---|---|---|---|---|---|
| ................................................CACATCCGGAAATCTAGTAGA........................................................................................ | 21 | 0 | 1 | 5.00 | 5 | 2 | 3 | 0 |
| ..............................................TACACATCCGGAAATCTAGTAGA........................................................................................ | 23 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 |
| ...............................................ACACATCCGGAAATCTAGTAGA........................................................................................ | 22 | 0 | 1 | 3.00 | 3 | 1 | 2 | 0 |
| .............................................ATACACATCCGGAAATCTAGTAG......................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ...............................................CCACATCCGGAAATCTAGTAGA........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..............................................TACACATCCGGAAATCTAGTAG......................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ..................................................CATCCGGAAATCTAGTAGA........................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .............................................TTACACATCAGGAAATCTAGTAG......................................................................................... | 23 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 |
Generated: 09/01/2015 at 12:08 PM