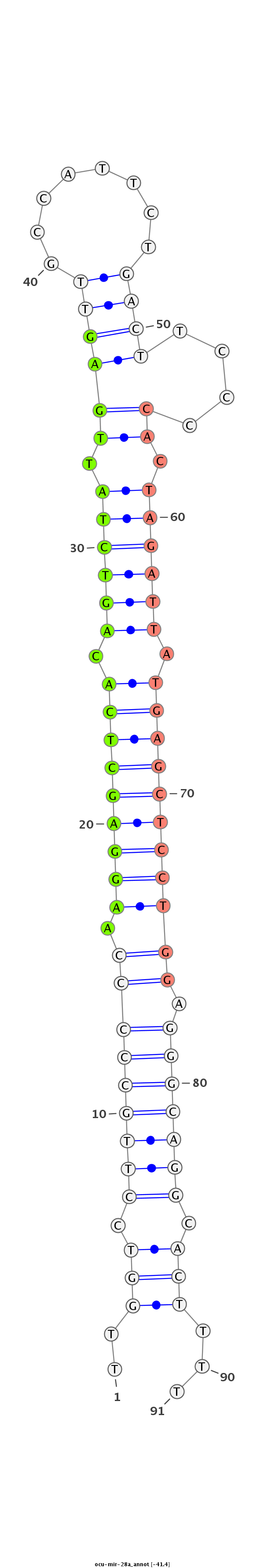
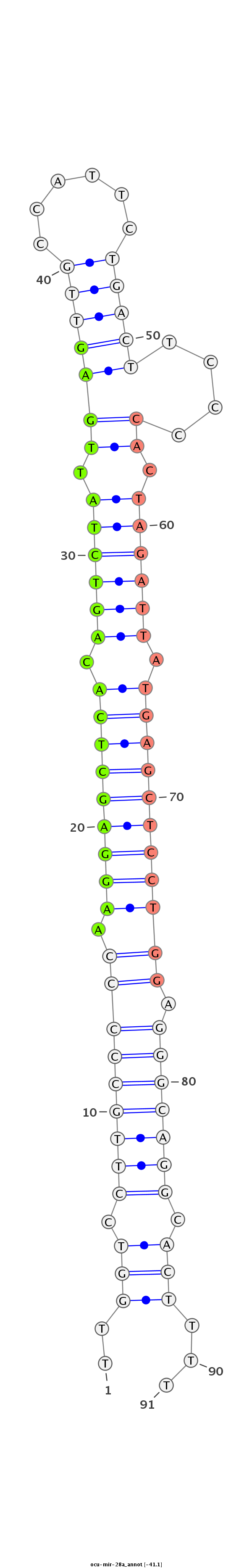
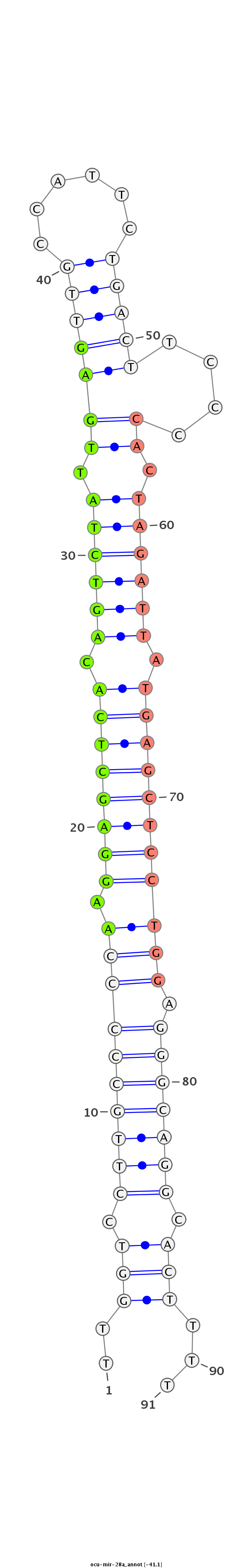
| ...............................................................AAGGAGCTCACAGTCTATTGAG..................................................................................................... |
22 |
0 |
1 |
110.00 |
110 |
109 |
1 |
| ...............................................................AAGGAGCTCACAGTCTATTGA...................................................................................................... |
21 |
0 |
1 |
25.00 |
25 |
16 |
9 |
| ...............................................................AAGGAGCTCACAGTCTATTG....................................................................................................... |
20 |
0 |
1 |
24.00 |
24 |
13 |
11 |
| ...............................................................AAGGAGCTCACAGTCTATTGAGT.................................................................................................... |
23 |
0 |
1 |
14.00 |
14 |
14 |
0 |
| ...............................................................AAGGAGCTCACAGTCTATT........................................................................................................ |
19 |
0 |
1 |
9.00 |
9 |
6 |
3 |
| ...............................................................AAGGACCTCACAGTCTATTGAG..................................................................................................... |
22 |
1 |
1 |
3.00 |
3 |
3 |
0 |
| ..................................................................GAGCTCACAGTCTATTGAGT.................................................................................................... |
20 |
0 |
1 |
2.00 |
2 |
2 |
0 |
| .......................................................................................................CACTAGATTATGAGCTCCTGG.............................................................. |
21 |
0 |
1 |
2.00 |
2 |
0 |
2 |
| .......................................................................................................CACTAGATTATGAGCTCCTGGT............................................................. |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
| ..............................................................CAAGGAGCTCACAGTCTAT......................................................................................................... |
19 |
0 |
1 |
2.00 |
2 |
2 |
0 |
| ......................................................................................................CCACTAGATTATGAGCTCCTG............................................................... |
21 |
0 |
1 |
2.00 |
2 |
0 |
2 |
| ...............................................................AAGGAGCTCACAGTCTATTGAT..................................................................................................... |
22 |
1 |
2 |
1.50 |
3 |
3 |
0 |
| ......................................................................................................CCACTAGATTATGAGCTCCTGG.............................................................. |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
| ................................................................AGGAGCTCACAGTCTATTGAG..................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| .......................................................................................................CACTAGATTATGAGCTCCTG............................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
1 |
| ...................................................................AGCTCACAGTCTATTGAG..................................................................................................... |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
| .......................................................................................................CACTAGATTATGAGCTCCTGGA............................................................. |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
| ...............................................................AAGGAGCTCACAGTCTAT......................................................................................................... |
18 |
0 |
2 |
1.00 |
2 |
1 |
1 |
| ..............................................................AAAGGAGCTCACAGTCTATTGAG..................................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| ...............................................................AAGGCGCTCACAGTCTATTGAG..................................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
| .......................................................................................................CACTAGATTGTGAGCTCCTGG.............................................................. |
21 |
1 |
5 |
0.60 |
3 |
3 |
0 |
| ...............................................................AAGGAGCTCACAGTCTATTGAA..................................................................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
| .......................................................................................................CACTAGATTGTGAGCTCCTG............................................................... |
20 |
1 |
10 |
0.10 |
1 |
1 |
0 |