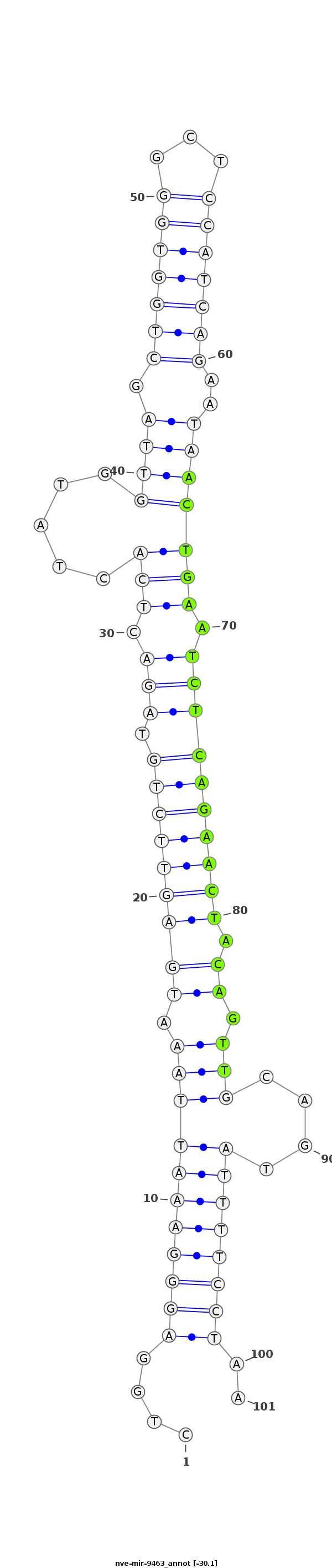
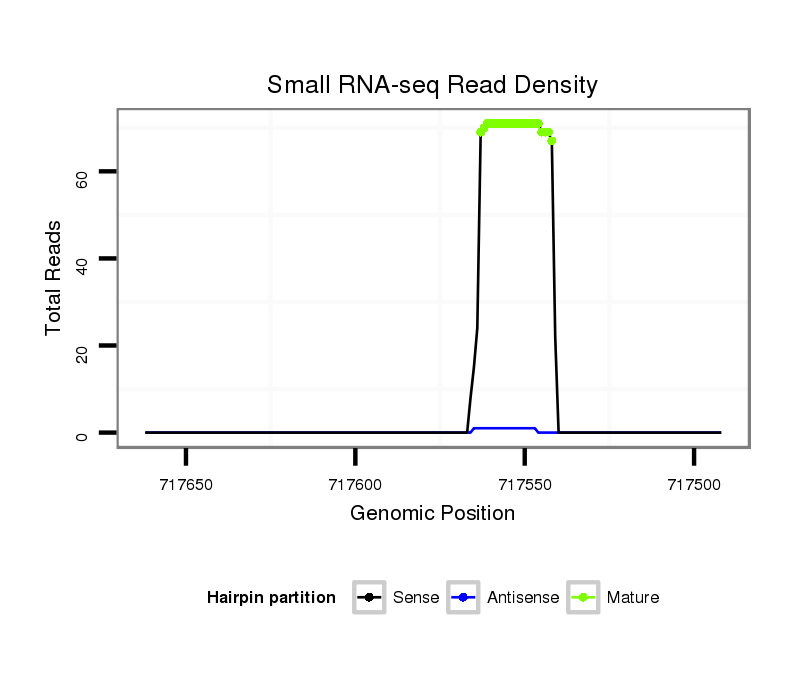
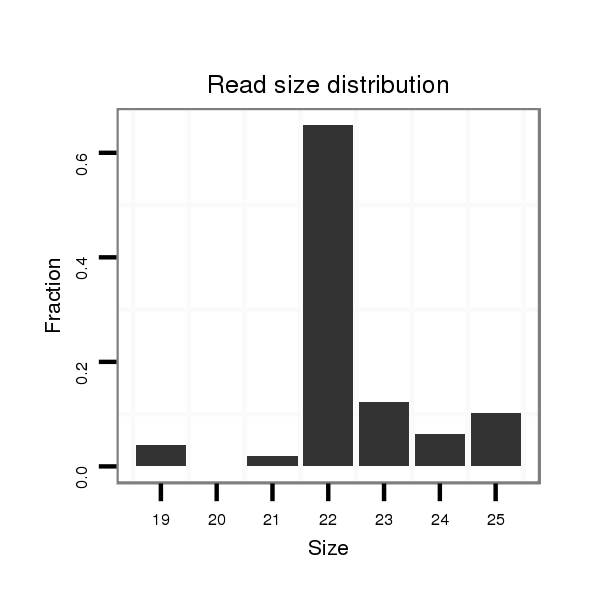
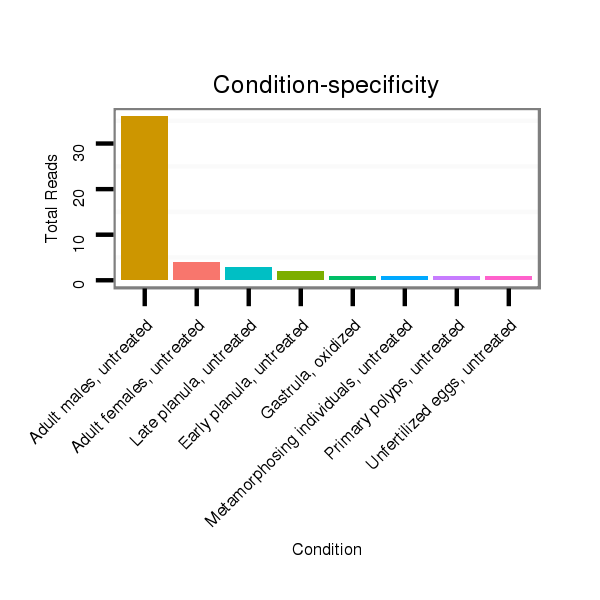
ID:nve-mir-9463 |
Coordinate:scaffold_94:717542-717612 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -30.1 | -29.8 | -29.8 |
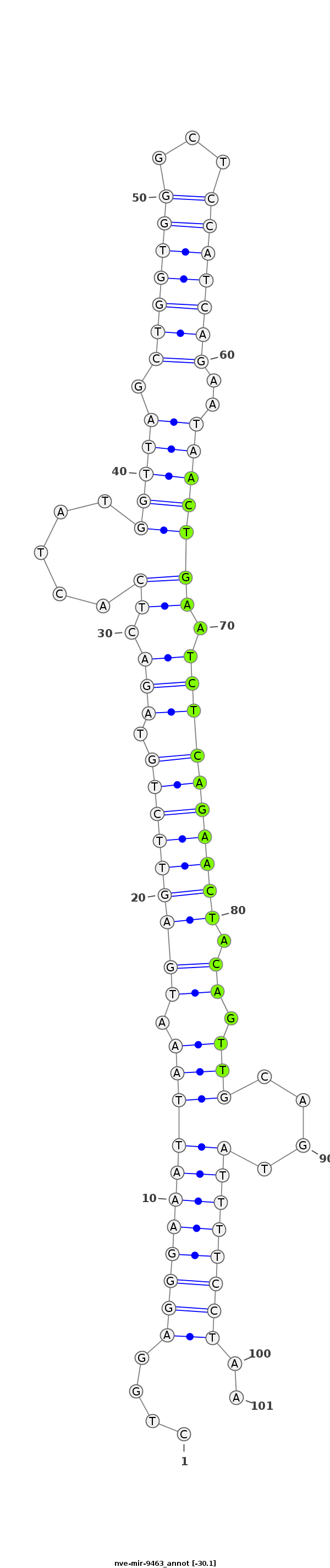 |
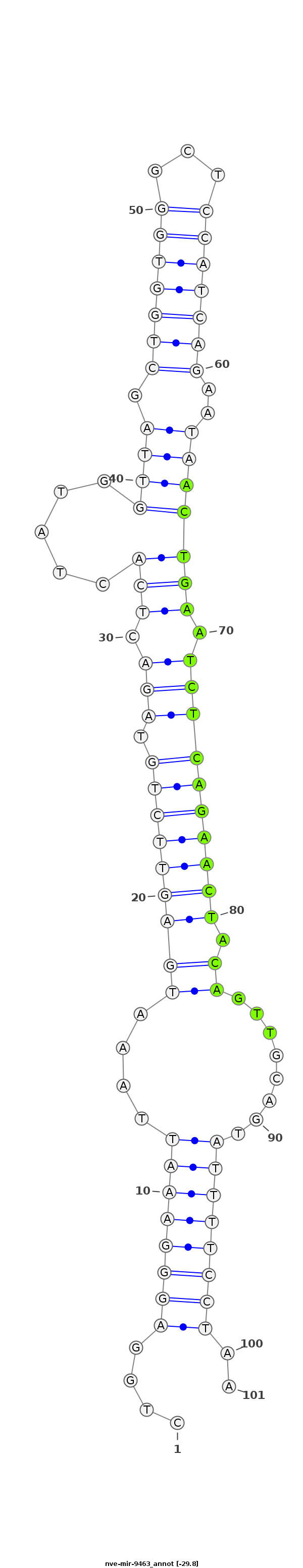 |
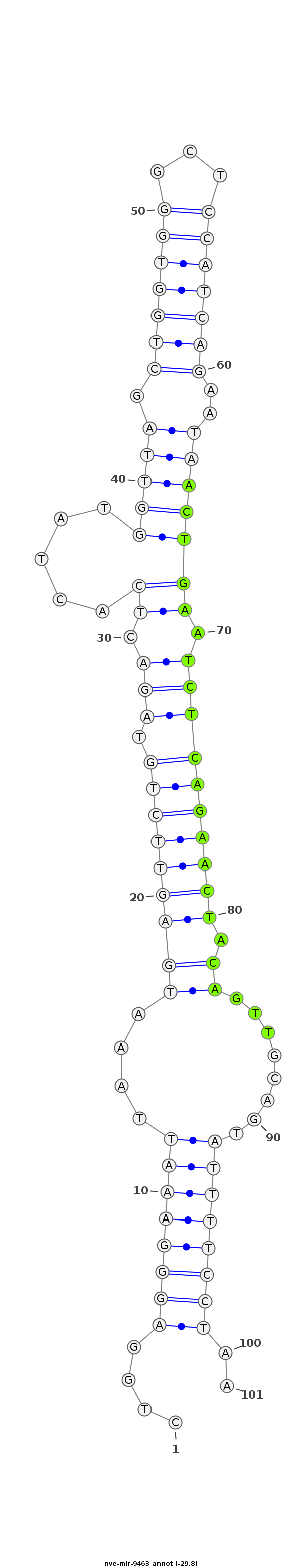 |
|
CATTACTGTCTACCTCTGTGGCAGTTGTGTAATACCTGGAGGGAAATTAAATGAGTTCTGTAGACTCACTATGGTTAGCTGGTGGGCTCCATCAGAATAACTGAATCTCAGAACTACAGTTGCAGTATTTTCCTAAAAGAACGCAAATGCCAAAACCGATAGGAAAACATA
***********************************....(((((((((((.(((((((((.(((.(((.....((((.(((((((...)))))))..))))))).)))))))))).)).)))....))))))))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039754 "Adult males, untreated" |
SRR039758 "Early planula, untreated" |
SRR039760 "Late planula, untreated" |
SRR039731 "Adult females, untreated" |
SRR039764 "Primary polyps, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039726 "Blastula, untreated" |
SRR039727 "Unfertilized eggs, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039757 "Gastrula, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................ACTGAATCTCAGAACTACAGTT.................................................. | 22 | 0 | 1 | 32.00 | 32 | 30 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCTCAGAACTACAGTTG................................................. | 23 | 0 | 1 | 12.00 | 12 | 10 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AACTGAATCTCAGAACTACAGTT.................................................. | 23 | 0 | 1 | 5.00 | 5 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................ATAACTGAATCTCAGAACTACAGTT.................................................. | 25 | 0 | 1 | 5.00 | 5 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TAACTGAATCTCAGAACTACAGTTG................................................. | 25 | 0 | 1 | 3.00 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................ATAACTGAATCTCAGAACTACAGTTG................................................. | 26 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TAACTGAATCTCAGAACTACAGTT.................................................. | 24 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................AACTGAATCTCAGAACTAC...................................................... | 19 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................AACTGAATCTCAGAACTACAGTTG................................................. | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................ATAACTGAATCTCAGAACTACAGCT.................................................. | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AACGGAAGCTCAGAACTACAGTTG................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TAACTGAATCTCAGAACTACAGT................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AACTGAAGCTCAGAACTACAGTT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AACTGAATCTCATAACTACATTT.................................................. | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................AATGAATCTCAGAACTACAGTT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCGCAGAACTACAGTT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCTCAGGACTACAGTTG................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AATCTCAGAACTACAGTTT................................................. | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGACTGAATCTCAGAACTGCAGTTG................................................. | 25 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCTCAGAACTACAGTTGCAGTTTTAT........................................ | 32 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATATCAGAACTATAGTT.................................................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AACTGAATCTCAGAACTACAGAT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TAGAATAACTGAATCTCAGAACTACAGTT.................................................. | 29 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CTGAATCTCAGAACTACAGTTG................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TGAATCTCAGAACTACAGTTG................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................AACGGAATCTCAGAACTACAGTTGA................................................ | 25 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCTCAGAACTACAGAT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCTCAGAACTACAGT................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCTCAGCACTACAGTT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGAATCTAAGAACTACAGTT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TAACTGAGTATCAGAACTACAGTT.................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................ACTGGATCTCAGAACTACAGTTG................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTAATGACAGATGGAGACACCGTCAACACATTATGGACCTCCCTTTAATTTACTCAAGACATCTGAGTGATACCAATCGACCACCCGAGGTAGTCTTATTGACTTAGAGTCTTGATGTCAACGTCATAAAAGGATTTTCTTGCGTTTACGGTTTTGGCTATCCTTTTGTAT
***********************************....(((((((((((.(((((((((.(((.(((.....((((.(((((((...)))))))..))))))).)))))))))).)).)))....))))))))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039731 "Adult females, untreated" |
|---|---|---|---|---|---|---|
| .................................................................................................ATTGACTTAGAGTCTTGAT....................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 08/21/2014 at 10:51 AM