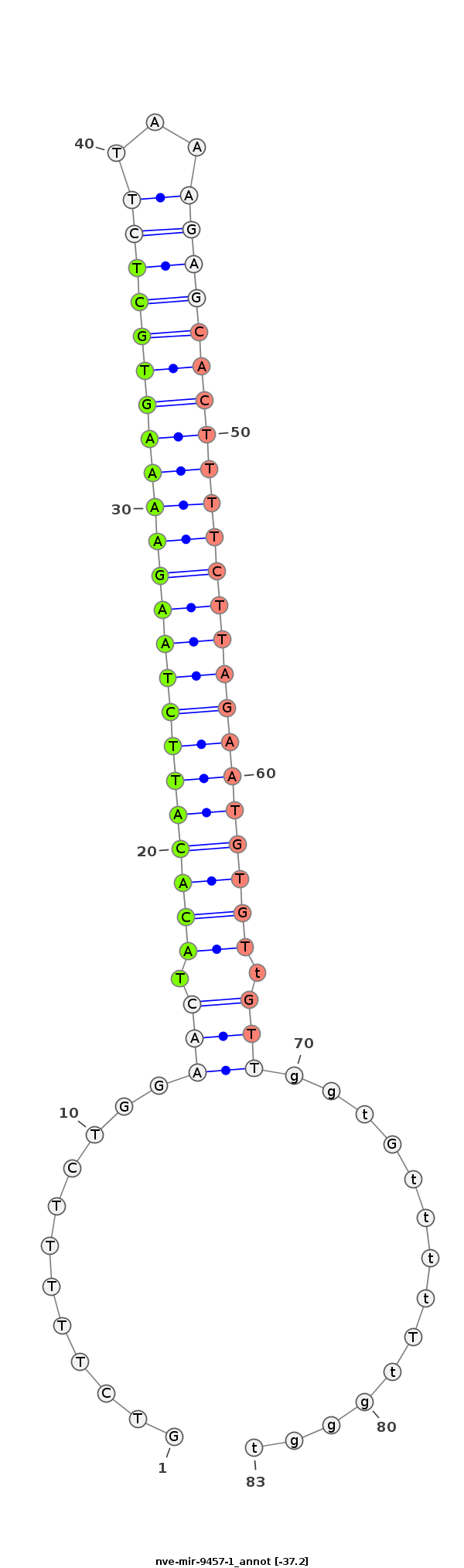
ID:nve-mir-9457-1 |
Coordinate:scaffold_1:1934606-1934673 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
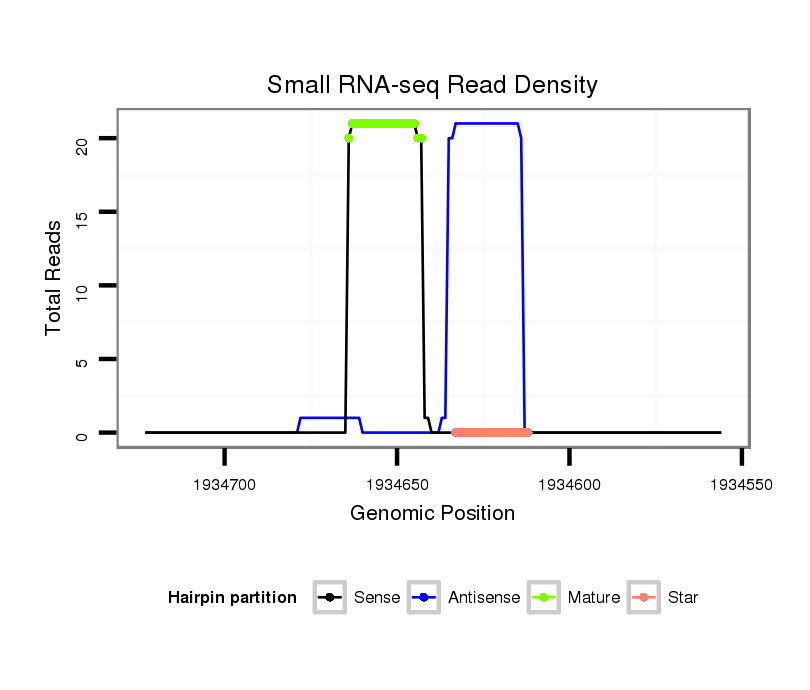
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
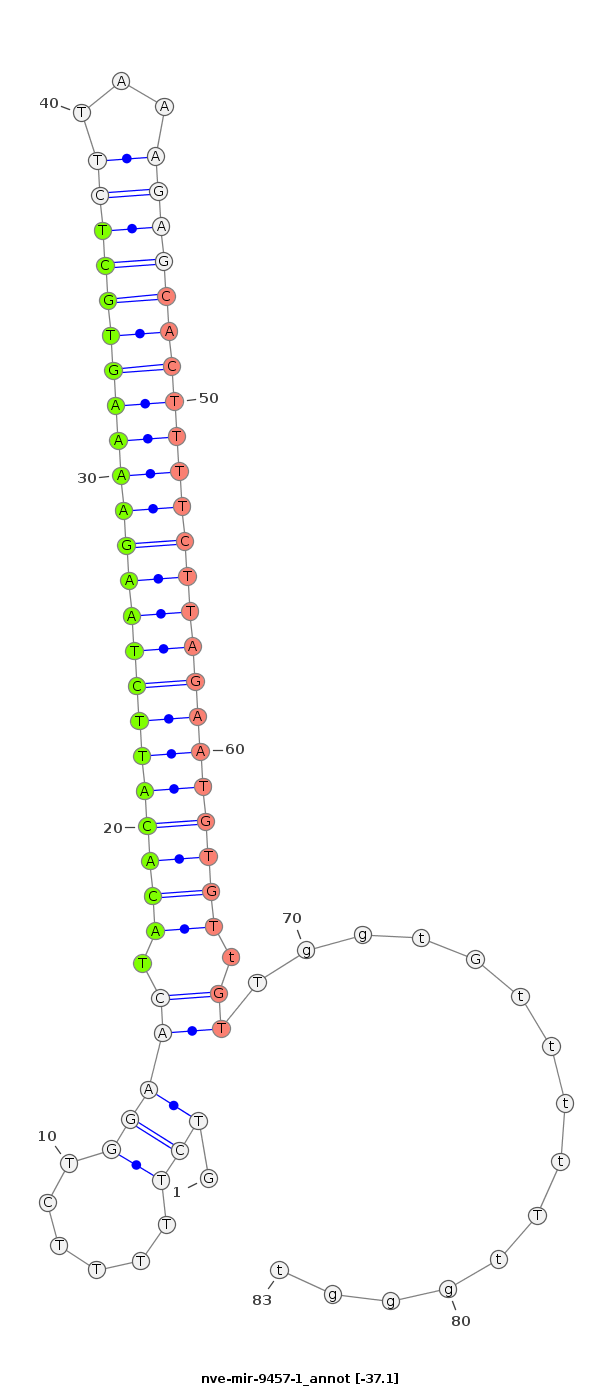
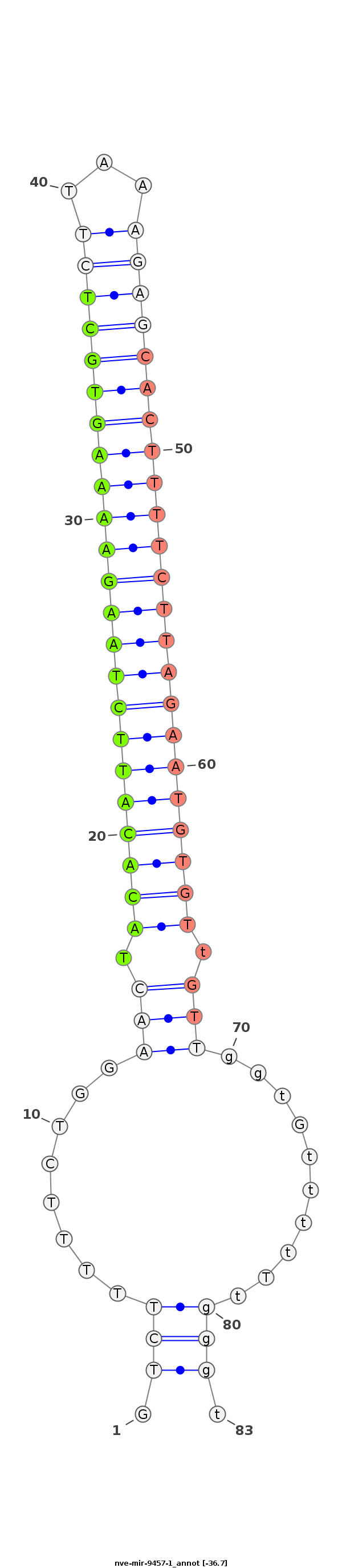
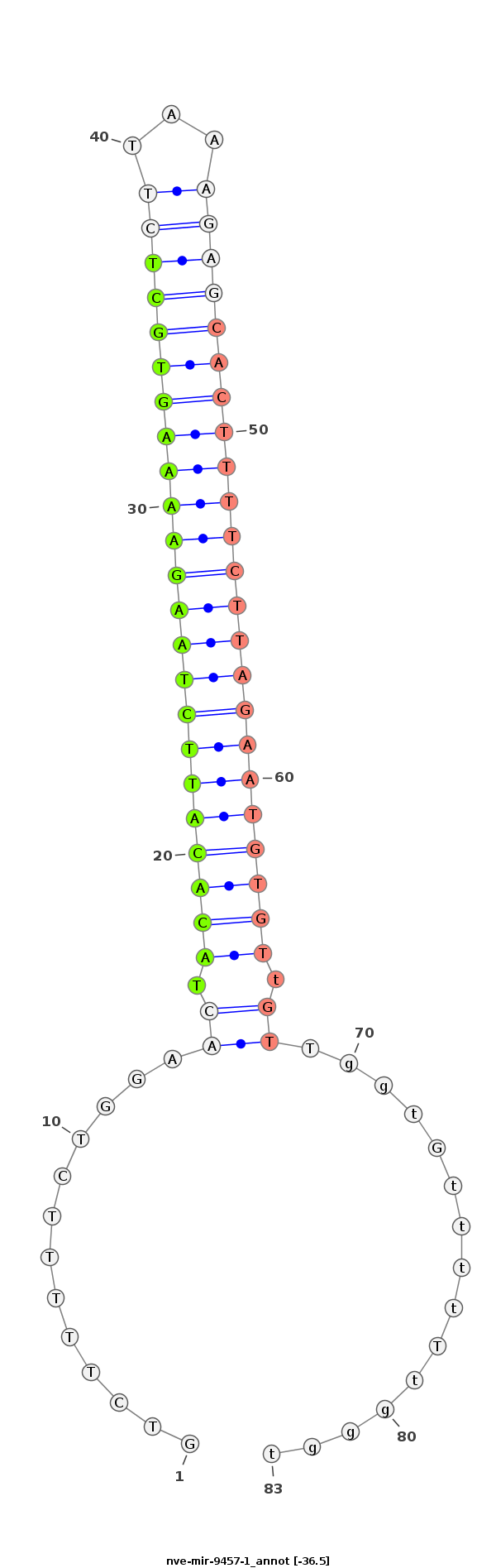
| -37.1 | -36.7 | -36.5 |
 |
 |
 |
| mature | star |
|
AGCAGCTGGCACAAGAAGACTATCCCCTTTGGCTGCTCCCTGATGTCTTTTTCTGGAACTACACATTCTAAGAAAAGTGCTCTTAAAGAGCACTTTTCTTAGAATGTGTtGTTggtGttttTtgggtggggggggggggGggtTGGtGGGGGTtGtTtTGtggggtgg
********************************************............(((.(((((((((((((((((((((((...))))))))))))))))))))))).)))..............***************************************** |
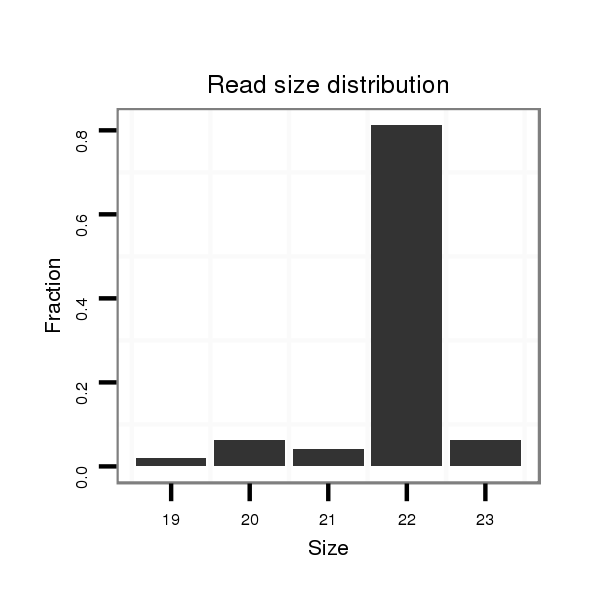
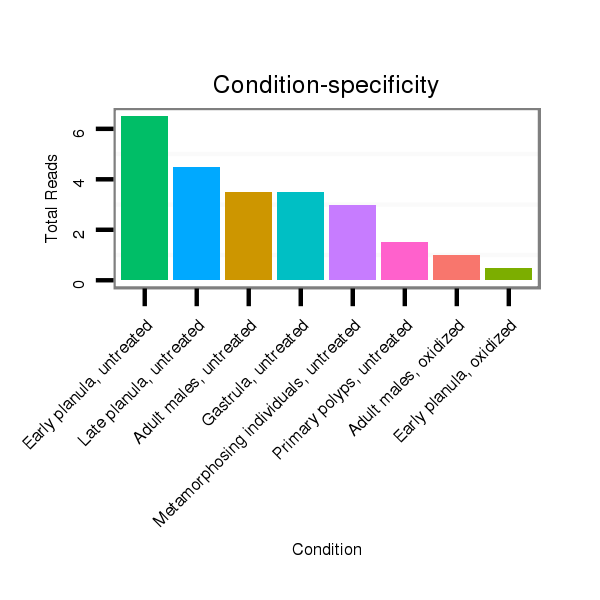
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039758 "Early planula, untreated" |
SRR039760 "Late planula, untreated" |
SRR039754 "Adult males, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039764 "Primary polyps, untreated" |
SRR039755 "Adult males, oxidized" |
SRR039759 "Early planula, oxidized" |
SRR039765 "Primary polyps, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TACACATTCTAAGAAAAGTGCT....................................................................................... | 22 | 0 | 2 | 19.00 | 38 | 11 | 8 | 3 | 6 | 6 | 2 | 2 | 0 | 0 |
| ............................................................ACACATTCTAAGAAAAGTGCTCT..................................................................................... | 23 | 0 | 2 | 1.00 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................TACACATTCTAAGAAAAGTG......................................................................................... | 20 | 0 | 2 | 1.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CATTCTAAGAAAAGTGCTCT..................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................TACACATTCTAAGAAAAGT.......................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACAGATTCTAAGAAAAGTGCT....................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTCAAAGAAAAGCGCT....................................................................................... | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................ACACATTCTAAGAAAAGTGCT....................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTCTAAGGAAAGTGCT....................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTCTAAGAAAAGTGCTTT..................................................................................... | 24 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................CACTTTTCTTAGAATGTGTAGT........................................................ | 22 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TAAACATTCGAAGAAAAGTGCT....................................................................................... | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTCTAAGAAAAGTGCTC...................................................................................... | 23 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTCTAAGAAAAATGCT....................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTCTAATAAAATTGCT....................................................................................... | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTCTAAGAAAAGTGC........................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TACACATTTTAAGAAAAGAGCG....................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................CTTAGAATCTGTAGTCCC..................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TCGTCGACCGTGTTCTTCTGATAGGGGAAACCGACGAGGGACTACAGAAAAAGACCTTGATGTGTAAGATTCTTTTCACGAGAATTTCTCGTGAAAAGAATCTTACACAtCAAggtCttttAtgggtggggggggggggCggtACCtCCCCCAtCtAtACtggggtgg
*****************************************............(((.(((((((((((((((((((((((...))))))))))))))))))))))).)))..............******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039758 "Early planula, untreated" |
SRR039760 "Late planula, untreated" |
SRR039754 "Adult males, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039764 "Primary polyps, untreated" |
SRR039731 "Adult females, untreated" |
SRR039755 "Adult males, oxidized" |
SRR039759 "Early planula, oxidized" |
SRR039765 "Primary polyps, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................TCGTGAAAAGAATCTTACACAT.......................................................... | 22 | 0 | 2 | 19.00 | 38 | 11 | 8 | 3 | 6 | 6 | 2 | 0 | 2 | 0 | 0 |
| .............................................AGAAAAAGACCTTGATGT......................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................GTGAAAAGAATCTTACACAT.......................................................... | 20 | 0 | 2 | 1.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TCTCGTGAAAAGAATCTTACACA........................................................... | 23 | 0 | 2 | 1.00 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TCGTTAAAATAATCTTACACAT.......................................................... | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TGAAAAGAATCTTACACAT.......................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCGCGAAAAGAAACTTACACAT.......................................................... | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCGTGAAAAGAAGCTTACAAAT.......................................................... | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGATGTGTAAGATTCTTTTCAC......................................................................................... | 22 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCGTGAAAAGAATCTTACACA........................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CGTGAAAAGAATCTTACACAT.......................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................CTCGTGAAAAGAATCTTACACAT.......................................................... | 23 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCGTAAAAAGAATCTTACACAT.......................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TCGTGAAAAGAATCTTAGACAT.......................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TTTCGTGAAAAGAATCTTACACAT.......................................................... | 24 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TCTCGTGAAAAGAATCTTAC.............................................................. | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TCGTGAAAGGAATCTTACACAT.......................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................GCGAGAAAAGAATTTTACACAT.......................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CCCTGATGTCTAAGATTC................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................ATCGACTACAGAAAAAGTCCT............................................................................................................... | 21 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................GCGTCGGGGGGAGGGGCGGTA........................ | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Generated: 08/21/2014 at 09:13 AM