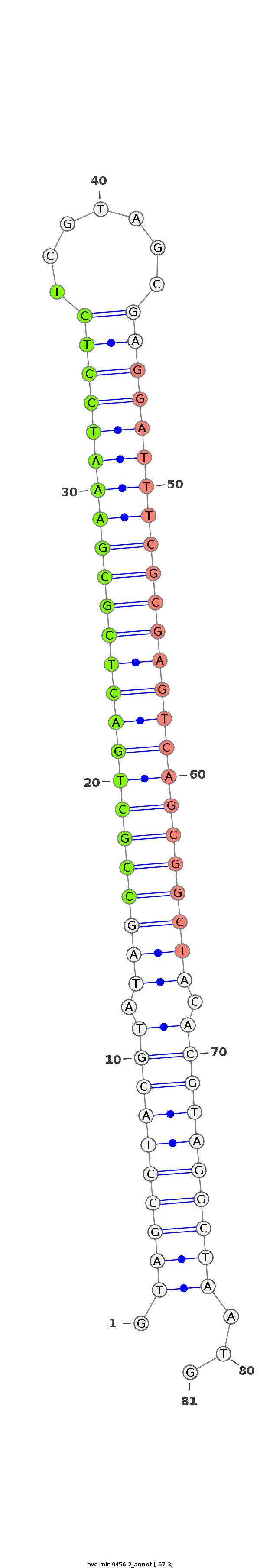
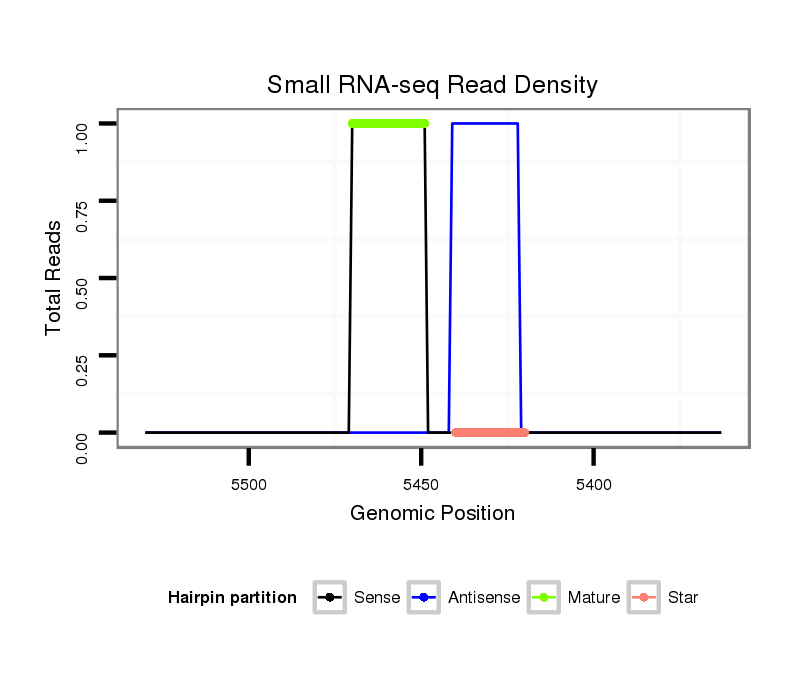
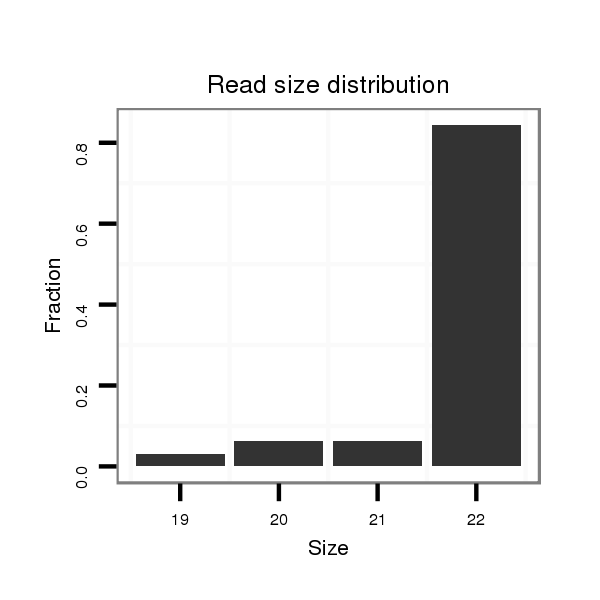

ID:nve-mir-9456-2 |
Coordinate:scaffold_699:5413-5480 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
AAGCTATTATAGTCCTTCCAAGTCCTTCTAGGTTGTTATGGAAAAGTAGCCTACGTATAGCCGCTGACTCGCGAAATCCTCTCGTAGCGAGGATTTCGCGAGTCAGCGGCTACACGTAGGCTAATGAAAGCTTCGATCCTAAACATGTGCCACTGAACGAATCAGATA
*********************************************.((((((((((.((((((((((((((((((((((((.......)))))))))))))))))))))))).))))))))))...****************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039756 "Gastrula, untreated" |
SRR039758 "Early planula, untreated" |
SRR039760 "Late planula, untreated" |
SRR039764 "Primary polyps, untreated" |
SRR039754 "Adult males, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039761 "Late planula, oxidized" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039765 "Primary polyps, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................CCGCTGACTCGCGAAATCCTCT...................................................................................... | 22 | 0 | 20 | 1.30 | 26 | 6 | 6 | 2 | 5 | 3 | 2 | 1 | 0 | 1 |
| ............................................................CCGCTGACTCGCGAAATCCTCA...................................................................................... | 22 | 1 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CCGCTGACTCGCGAAATCCT........................................................................................ | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..........................................................................................GGATTTCGCGAGTCAGCGGCT......................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CCGCTGACTCGAGAAATCCGCT...................................................................................... | 22 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CCGCTGACTCGCGAAATCTTCT...................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................AGGATTTCGCGAGTCAGCGGCT......................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CTGACTCGCGAAATCCTCTCT.................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CCGCTGACTCGCGAAATCCTA....................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................TAGCCGCTGACTCGCGAAATC.......................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CCGCTGACTCGCGACATCCT........................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CCGCTGACTCGCGAAATCC......................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................AGGCTTTCGCGCGTCAGCGGCT......................................................... | 22 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................AGGATTTCGCGAGTCAGCGGT.......................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................TAGGTTGTTATGGAG............................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................CCGCCGACTCGCGAAATCCT........................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTCGATAATATCAGGAAGGTTCAGGAAGATCCAACAATACCTTTTCATCGGATGCATATCGGCGACTGAGCGCTTTAGGAGAGCATCGCTCCTAAAGCGCTCAGTCGCCGATGTGCATCCGATTACTTTCGAAGCTAGGATTTGTACACGGTGACTTGCTTAGTCTAT
******************************************.((((((((((.((((((((((((((((((((((((.......)))))))))))))))))))))))).))))))))))...********************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039764 "Primary polyps, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039754 "Adult males, untreated" |
SRR039758 "Early planula, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039760 "Late planula, untreated" |
SRR039731 "Adult females, untreated" |
SRR039765 "Primary polyps, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................TCCTAAAGCGCTCAGTCGCC........................................................... | 20 | 0 | 20 | 1.20 | 24 | 10 | 0 | 4 | 6 | 2 | 2 | 0 | 0 |
| .................................................GGATGCATATCGGCGACTGAGC................................................................................................. | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TGCATATCGGCGACTGAGC................................................................................................. | 19 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TTTCATATCGGCGACTGAGC................................................................................................. | 20 | 2 | 12 | 0.17 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................GCGCTCAGTCGCCGATGTGCAT.................................................. | 22 | 0 | 20 | 0.15 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................TTTCGGCGACTGAGCGCTCTAGGA........................................................................................ | 24 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................TATTTCGGCGACTGAGCGCTCTAGG......................................................................................... | 25 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................CCTAAAGCGCTCAGTCGCC........................................................... | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................CTCCTAAAGCGCTCAGTCGC............................................................ | 20 | 0 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................TACTTTCGGAGCTAGGACTT......................... | 20 | 2 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCGGCGACTGAGCGCTTTAGG......................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCTAAAGCGCTCATTCGCC........................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTCATAAAGCGCTCAGTCGCC........................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCTAAAGCGCTCAGGCGCC........................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCTACAGCGCTCAGTCGCC........................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................CGGCGACTGAGCGCTGTAGGA........................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................CTAAAGCGCTCAGTCGCC........................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................GCGCTCAGTCGCCGATGTGCA................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................ATCCTAAAGCGCTCAGTCGCC........................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTCCTAAAGCGCTCAGTCGCC........................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................TCTCTCCTAAAGCGCTCAGTC.............................................................. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................ACTCCTAAAGCGCTCAGTCGCC........................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCGGCGACTGCGCGCTTTCGGA........................................................................................ | 22 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................CTCCTAAAGCGCTCGGTCGCC........................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTCCTAAATCTCTCAGTCGCC........................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................ACTGAGCGCTTTAGGAGA...................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................GAGCTCAGTCGCCGGTGTGCAT.................................................. | 22 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................TTCCTAAAGCGCTCAGTCGCC........................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCGGCGACTGAGCGCTTTAGGA........................................................................................ | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................AGCGCTCAGTCGCCGATGTGCAT.................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCTAATGCGCTTAGTCGCC........................................................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Generated: 08/21/2014 at 10:46 AM