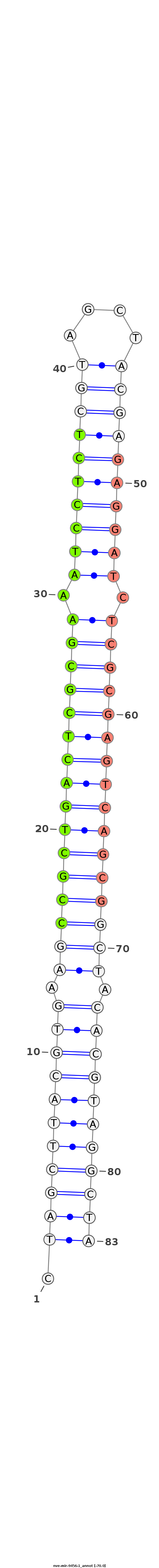

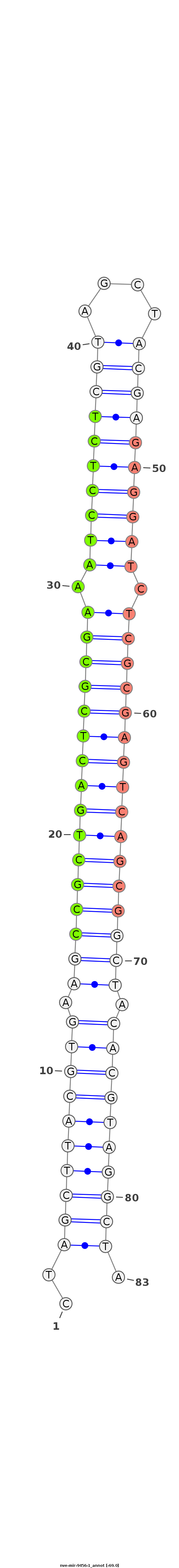
ID:nve-mir-9456-1 |
Coordinate:scaffold_148:42285-42352 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
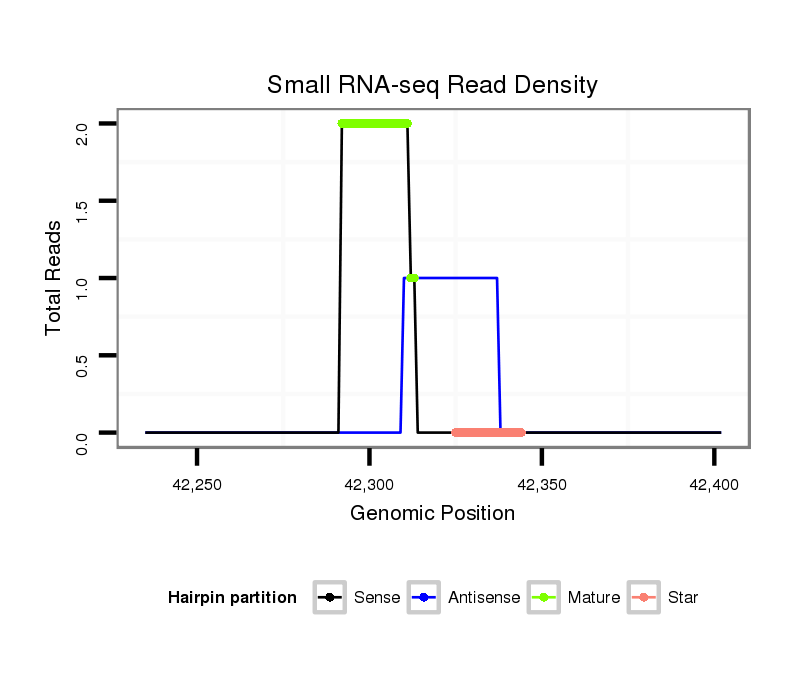
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -69.0 |
 |
|
GCTTGTTGTTGTTTAAAGTGTCCCTGTAAGCCAAAAGGCGACCTAGCTTACGTGAAGCCGCTGACTCGCGAAATCCTCTCGTAGCTACGAGAGGATCTCGCGAGTCAGCGGCTACACGTAGGCTAAAGGCGAcctagcaaagtatcaagtgtgagaaagcatctattt
******************************************.(((((((((((.((((((((((((((((.((((((((((....)))))))))).)))))))))))))))).)))))))))))******************************************* |
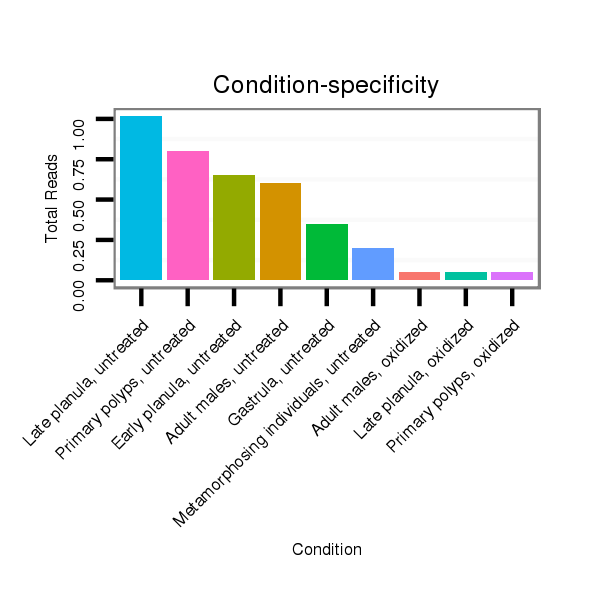
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039764 "Primary polyps, untreated" |
SRR039758 "Early planula, untreated" |
SRR039760 "Late planula, untreated" |
SRR039754 "Adult males, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039765 "Primary polyps, oxidized" |
SRR039761 "Late planula, oxidized" |
SRR039755 "Adult males, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................CCGCTGACTCGCGAAATCCTCT......................................................................................... | 22 | 0 | 20 | 1.20 | 24 | 5 | 6 | 2 | 0 | 6 | 2 | 0 | 1 | 1 | 1 |
| ...........................................................................................AGGATCTCGCGAGTCAGCGGCTT...................................................... | 23 | 1 | 7 | 1.14 | 8 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | 1 | 0 |
| .........................................................CCGCTGACTCGCGAAATCCT........................................................................................... | 20 | 0 | 20 | 1.10 | 22 | 10 | 6 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GAGGATCTCGCGAGTCAGCG.......................................................... | 20 | 0 | 5 | 0.40 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AGGATCTCGCGAGTCAGCGGT........................................................ | 21 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................AGGATCTCGCGAGTCAGCGGC........................................................ | 21 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GAGGATCTCGCGAGTCAGCGGC........................................................ | 22 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GAGGATCTCGCGAGTCAG............................................................ | 18 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AGGATCTCGCGAGTCAGCGGAT....................................................... | 22 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................AGAGGATCTCGCGAGTCA............................................................. | 18 | 0 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AGGATCTCGCGAGTCAGT........................................................... | 18 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GGGATCTCGCGAGTCAGCGG......................................................... | 20 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AGGATCTCGCGAGTCAGCGGCTTT..................................................... | 24 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................GGATCTCGCGAGTCAGCGGCTTTAT................................................... | 25 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................CGCTGACTCGCGAAATCCTC.......................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGAAATCCTCA......................................................................................... | 22 | 1 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGACATCCT........................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................CGCTGACTCGCGAAATCCTCT......................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGAAATCCTC.......................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AGGATGTCGCGAGTCAGCGGC........................................................ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGAAATACTC.......................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGAAATCC............................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTTACTCGCGAAATCCT........................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGAAATC............................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGGCTCGCGAAATCCTC.......................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCTCTAAATCCTC.......................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCCGACTCGCGAAATCCT........................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCTCTGACTCGCGAAATCCGCT......................................................................................... | 22 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGAAATCTTCT......................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCGGACTCGCGAAATCCT........................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGATTCGCGTAATCCT........................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CCGCTGACTCGCGAAATCCTT.......................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CGAACAACAACAAATTTCACAGGGACATTCGGTTTTCCGCTGGATCGAATGCACTTCGGCGACTGAGCGCTTTAGGAGAGCATCGATGCTCTCCTAGAGCGCTCAGTCGCCGATGTGCATCCGATTTCCGCTGGtTgGTTTgtTtGTTgtgtgTgTTTgGTtGtTttt
*******************************************.(((((((((((.((((((((((((((((.((((((((((....)))))))))).)))))))))))))))).)))))))))))****************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039764 "Primary polyps, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039758 "Early planula, untreated" |
SRR039754 "Adult males, untreated" |
SRR039761 "Late planula, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................TTCGGCGACTGAGCGCTCTAGGA........................................................................................... | 23 | 1 | 7 | 1.14 | 8 | 0 | 2 | 2 | 1 | 1 | 1 | 1 |
| ...........................................................................GAGAGCATCGATGCTCTCCTAGAGCGCT................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TTTCGGCGACTGAGCGCTCTAGGA........................................................................................... | 24 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................TATTTCGGCGACTGAGCGCTCTAGG............................................................................................ | 25 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................TGGCGACTGAGCGCTTTAGGA........................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................GACTGAGCGCTTTAGGAGAG........................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................TCGGCGACTGAGCGCTTTAGG............................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GACTGAGCGCTTTCGGAGAG........................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 08/21/2014 at 09:33 AM