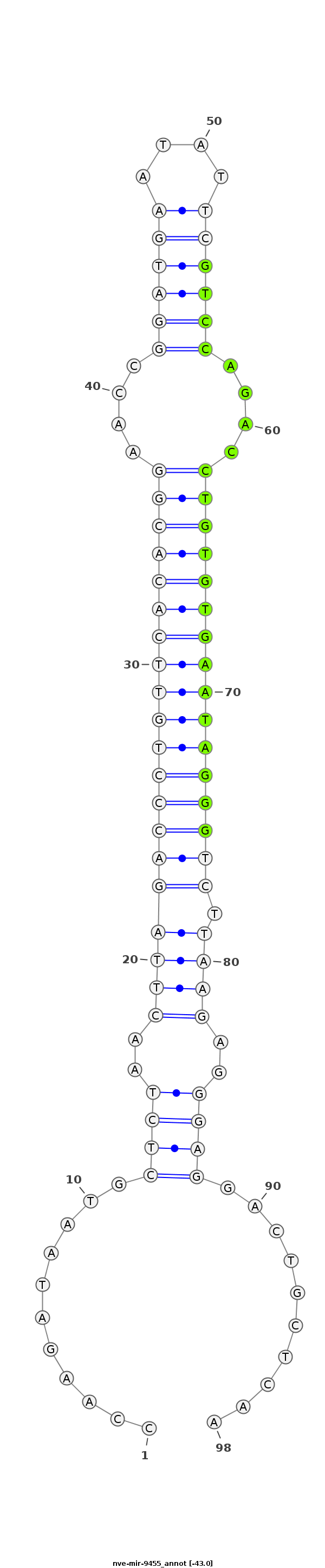
ID:nve-mir-9455 |
Coordinate:scaffold_18:243878-243945 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -43.0 | -42.8 | -42.6 |
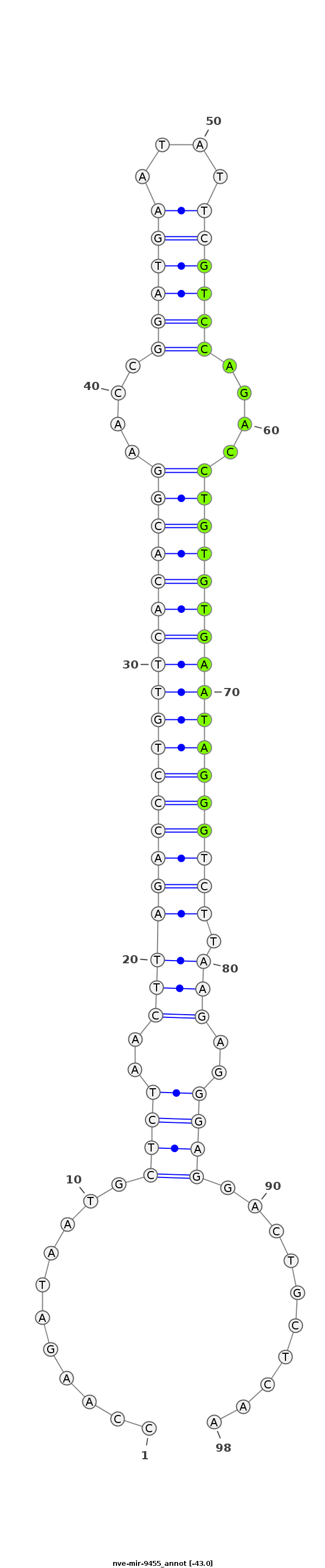 |
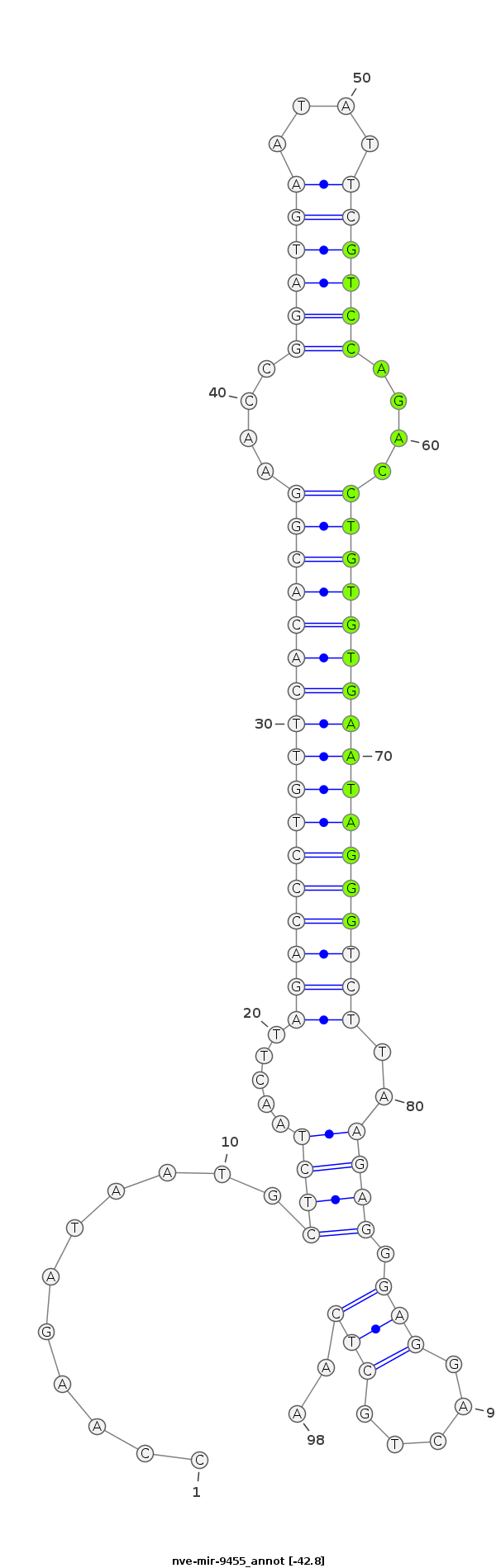 |
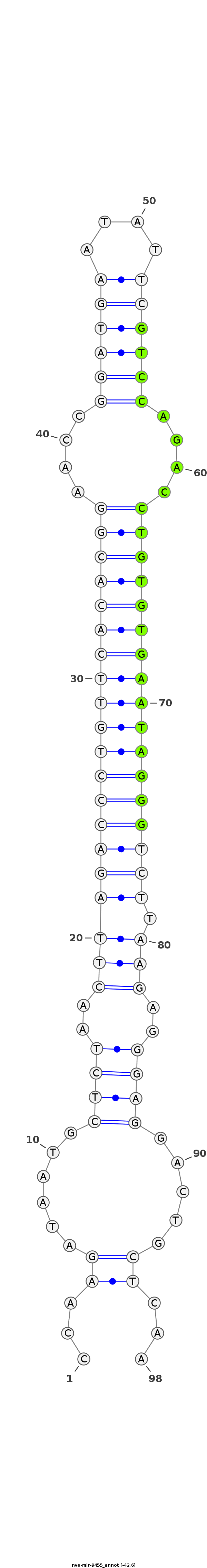 |
|
TAATACGTAAATTCTACCCCTCAGCATGTACCTGTCCAAGATAATGCTCTAACTTAGACCCTGTTCACACGGAACCGGATGAATATTCGTCCAGACCTGTGTGAATAGGGTCTTAAGAGGGAGGACTGCTCAAAGAAATAAAATTTAGAATCATTTTTAAAGGTACTG
***********************************...........((((..((((((((((((((((((((....((((((....))))))....)))))))))))))))).))))..))))..........*********************************** |
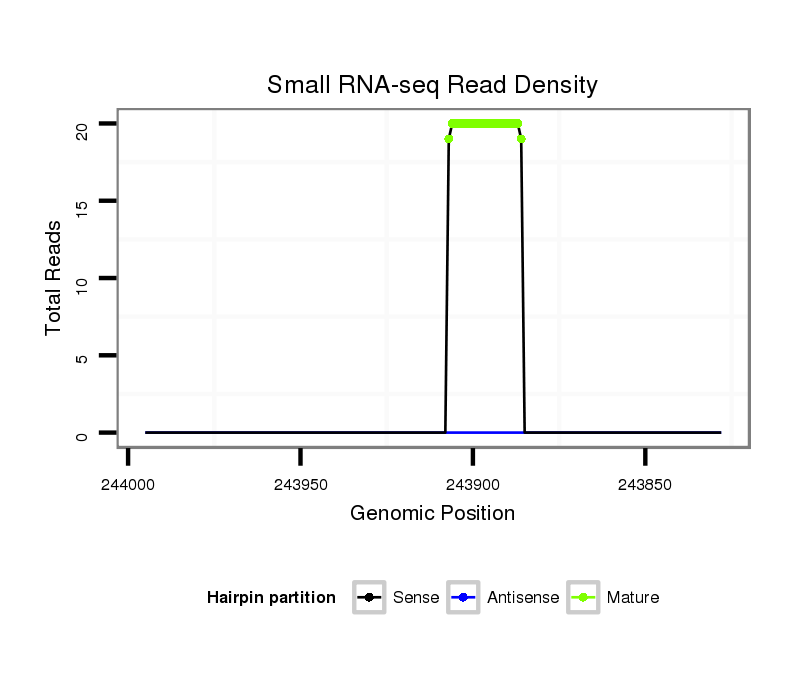
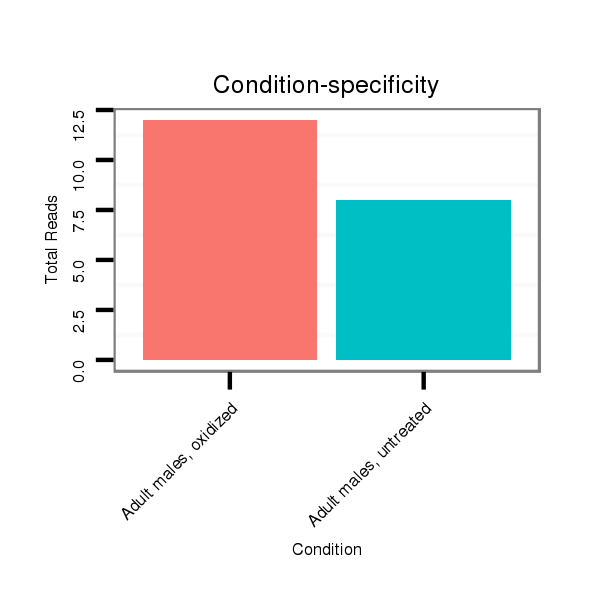
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039755 "Adult males, oxidized" |
SRR039754 "Adult males, untreated" |
SRR039731 "Adult females, untreated" |
SRR039765 "Primary polyps, oxidized" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039760 "Late planula, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|---|
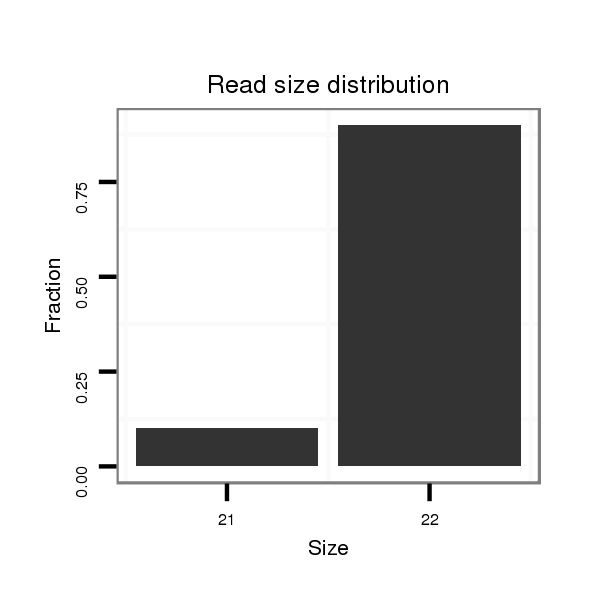
| ........................................................................................GTCCAGACCTGTGTGAATAGGG.......................................................... | 22 | 0 | 1 | 18.00 | 18 | 12 | 6 | 0 | 0 | 0 | 0 |
| .........................................TGATGTTCTAACTCAGACCC........................................................................................................... | 20 | 3 | 12 | 1.58 | 19 | 0 | 0 | 19 | 0 | 0 | 0 |
| ...........................................ATGTTCTAACTCAGACCC........................................................................................................... | 18 | 2 | 6 | 1.50 | 9 | 0 | 0 | 9 | 0 | 0 | 0 |
| ........................................................................................GTCCAGACATGAGTGAATAGGG.......................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................GTCCGGACCTGTGTGAATAGGG.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................GGCCAGACCTGTGTGAATAGGG.......................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TTCCAGACCTGCGTGAATAGGG.......................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................TGTTCACACGGAACCGGATGATT.................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCGGACCTGTGGGAATAGGGT......................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TCCAGACCTGTGTGAATAGGG.......................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................GTCCAGACCTGTGTGAATAGG........................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................GTCCAGACCTGTGTGACTATGG.......................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................TGATGTTCTAACGTAGACCC........................................................................................................... | 20 | 3 | 5 | 0.40 | 2 | 0 | 0 | 1 | 0 | 1 | 0 |
| ...........................................ATGCTCTAACGTAGGCCC........................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................TGATGGTCTAACTCAGACCC........................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................GATGTTCTAACGTAGACCC........................................................................................................... | 19 | 3 | 16 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ...........................................ATGTTCTAACTCAGACC............................................................................................................ | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ............................................TGCTCTAACTCAGACCCGT......................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................TTGATGTTCTAACTTAGACC............................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................TTAATGTTCTAACTCAGACC............................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................TGTGTGAATAAGGTCTTCGG................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATTATGCATTTAAGATGGGGAGTCGTACATGGACAGGTTCTATTACGAGATTGAATCTGGGACAAGTGTGCCTTGGCCTACTTATAAGCAGGTCTGGACACACTTATCCCAGAATTCTCCCTCCTGACGAGTTTCTTTATTTTAAATCTTAGTAAAAATTTCCATGAC
***********************************...........((((..((((((((((((((((((((....((((((....))))))....)))))))))))))))).))))..))))..........*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039755 "Adult males, oxidized" |
|---|---|---|---|---|---|---|
| .......ATGTAAGATGGGGAGT................................................................................................................................................. | 16 | 1 | 9 | 0.11 | 1 | 1 |
Generated: 08/21/2014 at 09:28 AM