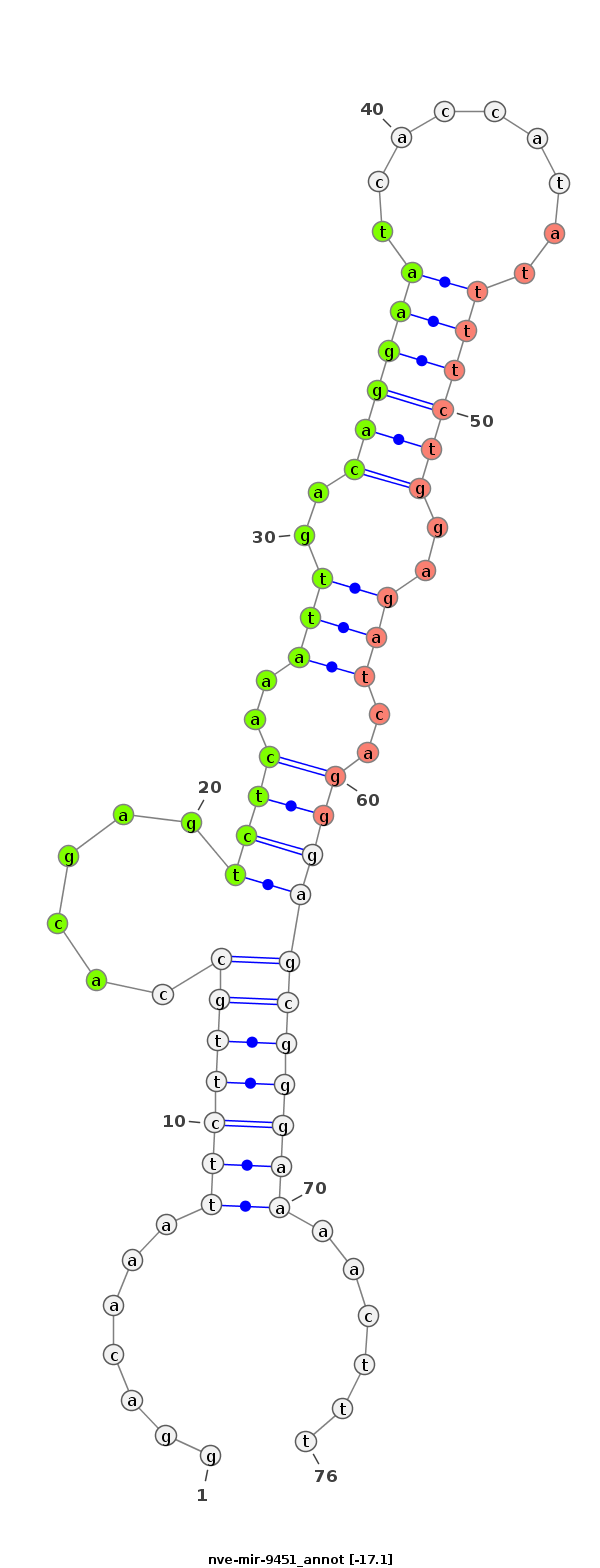
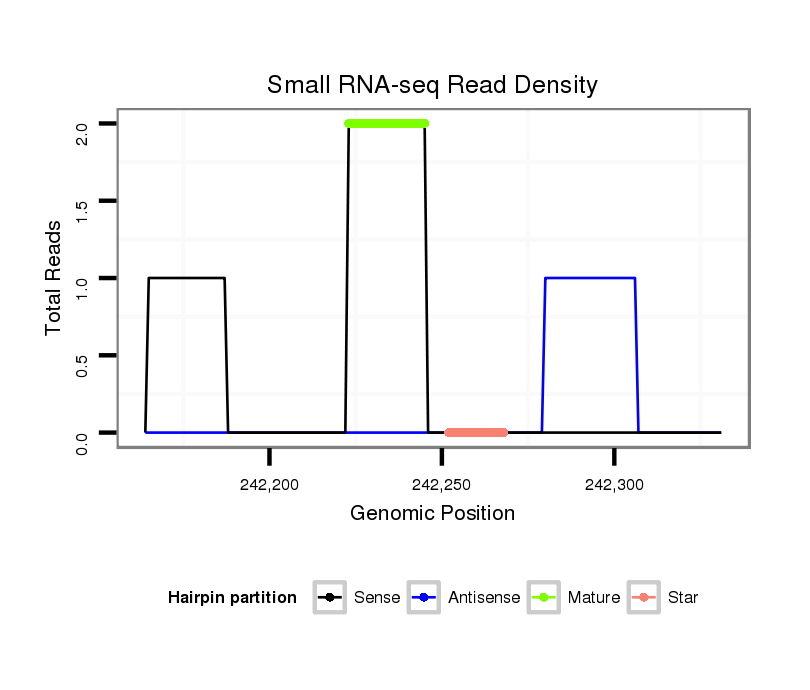
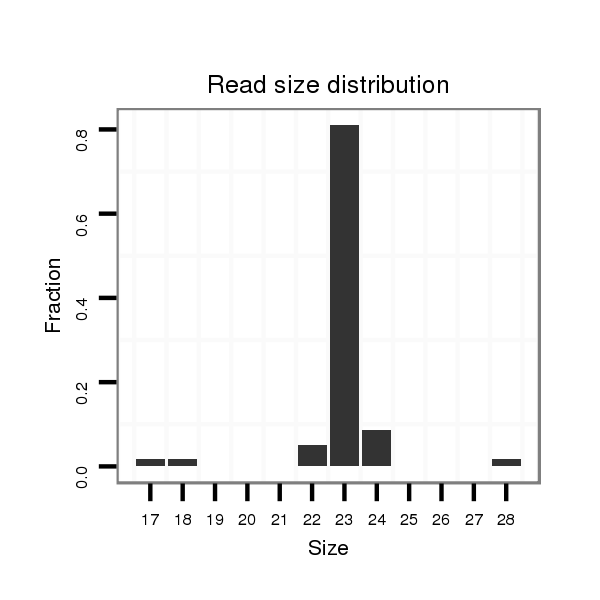
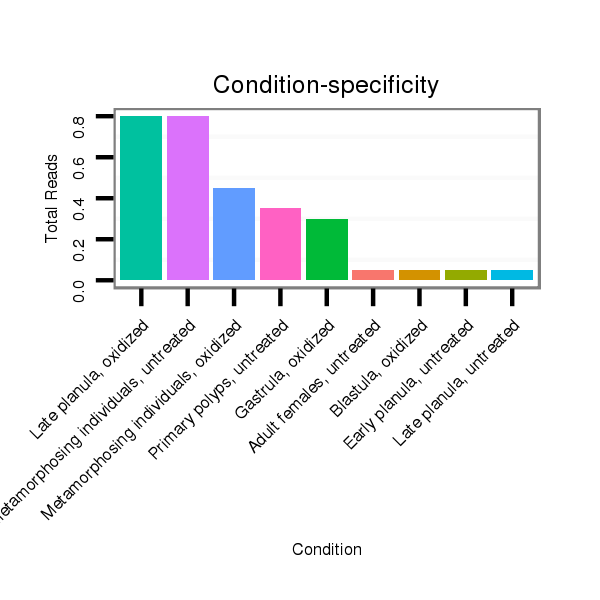
ID:nve-mir-9451 |
Coordinate:scaffold_319:242214-242281 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
aagattagtgagataaaaaattgaccaagctgtcaggtttttttggacaaattcttgccacgagtctcaaattgacaggaatcaccatattttctggagatcagggagcgggaaaactttagctcttataaatatgggcatcaatgaccatataaggcaatccttggg
********************************************.......(((((((......((((..(((..((((((.........))))))..)))..)))))))))))......************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039762 "Metamorphosing individuals, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039761 "Late planula, oxidized" |
SRR039757 "Gastrula, oxidized" |
SRR039764 "Primary polyps, untreated" |
SRR039758 "Early planula, untreated" |
SRR039729 "Blastula, oxidized" |
SRR039760 "Late planula, untreated" |
SRR039726 "Blastula, untreated" |
SRR039731 "Adult females, untreated" |
SRR039754 "Adult males, untreated" |
SRR039765 "Primary polyps, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................ACGAGTCTCAAATTGACAGGAAT...................................................................................... | 23 | 0 | 20 | 2.30 | 46 | 15 | 7 | 0 | 12 | 5 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .AGATTAGTGAGATAAAAAATTGA................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AGATTAGGGAGATAAAAAATTGAACAAAC.......................................................................................................................................... | 29 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TTTAGCTCTTATAAATATGTTCA............................ | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ACGAGTCTCAAATTGACCAGAATCACC.................................................................................. | 27 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ACGAGTCTCAAATTGACAGGAATC..................................................................................... | 24 | 0 | 20 | 0.25 | 5 | 1 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ACGAGTCTCAAATTGACAGGAA....................................................................................... | 22 | 0 | 20 | 0.15 | 3 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AGATTAGTGAGATAAAAA..................................................................................................................................................... | 18 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AGATTAGTGAGATAAAA...................................................................................................................................................... | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GAGTCTCAAATTGACAGGAATCA.................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................TTGGACAAATTCTTGCCACGGGTCTCAAA................................................................................................. | 29 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................ACGAGTCTCAACTTGACAGGAA....................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................AATATGGGCATCAATGACCATATAAGGC.......... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................ACGAGTCTCAAATTTACAGGAAT...................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TGGGCATCAATGACCATATAAGGCA......... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................ATTTTCTGGAGATCAGG............................................................... | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TGGGCATCAATGACCATATAAGGC.......... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................CAAGCTGTCAGGTTTTTTTGGACAAATTC.................................................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................ACGAGTCTCAAATTGACCGGAA....................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ACGAGTCTCAAATTGACA........................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................TGGAGATCAGGGAGCGGGAAAACTTTA............................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TATTTTCTGGAGATCAGGGAGCGGGAAA..................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ACGAGTCTCAAATTGCCAGGAAT...................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................AGGAGTCTCAAATTGACAGGAAT...................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .AGATTAGTGAGATAAAAACTTGACCAAGC.......................................................................................................................................... | 29 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ACGACTCTCAAATTGACAGGAAT...................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ATGAGTCTCAAATTGACAGGAAT...................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................CAAGCTGTCAGGTTTTTTTGGACAAATT................................................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................ACGAGTCTCAAATTGACATGAAT...................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTgTttTgtgTgTtTTTTTTttgTGGTTgGtgtGTggtttttttggTGTTTttGttgGGTGgTgtGtGTTTttgTGTggTTtGTGGTtTttttGtggTgTtGTgggTgGgggTTTTGtttTgGtGttTtTTTtTtgggGTtGTTtgTGGTtTtTTggGTTtGGttggg
************************************************.......(((((((......((((..(((..((((((.........))))))..)))..)))))))))))......******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039758 "Early planula, untreated" |
SRR039765 "Primary polyps, oxidized" |
SRR039764 "Primary polyps, untreated" |
SRR039757 "Gastrula, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039761 "Late planula, oxidized" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039760 "Late planula, untreated" |
SRR039727 "Unfertilized eggs, untreated" |
SRR039729 "Blastula, oxidized" |
SRR039731 "Adult females, untreated" |
SRR039754 "Adult males, untreated" |
SRR039759 "Early planula, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................GAAATCGAGAATATTTATACCCGTAGT......................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TGCTCAGAGTTTAACTGTCCTTCGTGGT................................................................................. | 28 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TGCTCAGAGTTTAACTGTCATTGGTGGT................................................................................. | 28 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CTGTTTAAGAACGGTGCTCAGAGTTT................................................................................................. | 26 | 0 | 20 | 0.35 | 7 | 0 | 0 | 0 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................ATATTTATACCCGTAGTAACTGGTATAT.............. | 28 | 1 | 6 | 0.33 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................TGTCCCTCGCCCTTTTTATATCGAGAAT........................................ | 28 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGTTTAAGAACGGTGCTCAGAGTTT................................................................................................. | 25 | 0 | 20 | 0.25 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GCTCAGAGCTTAACTGTCCTTGGTGGT................................................................................. | 27 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AATATTTATACCCGTAGTAACTGGTATAT.............. | 29 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................ATATTTATACCCGTAGTAACTGGTATATT............. | 29 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AATATTTATACCCTTAGTAACTGGTATAT.............. | 29 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TAATATTTATACCCTTAGTAACTGGTATAT.............. | 30 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GGTGCTCAGAGTTTAACTGTCCTTAGT.................................................................................... | 27 | 0 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................TCTCGTCCCTCGCCCTTTTGAAATCGAGAA......................................... | 30 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................TGTTTAAGAACGGTGCTCAGAGT................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AGTCCCTCGCCCTTTTGAAATCGAGA.......................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGTTTAAGCAGGGTGCTCAGCGTT.................................................................................................. | 24 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AGAACGGTGCTCAGAGTTTAACTGTCCT........................................................................................ | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................TACCCGTAGTTACTGGTAT................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CCGTTTAAGAACGGTGCTTAGAGTTT................................................................................................. | 26 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AGTACGTTGCTCAGAGTTTAACTGTCCT........................................................................................ | 28 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................GAACGGTGCTCAGAGTTTAACTGTCCTT....................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................AGAACGGTCCTCAGAGTTTAACTGTCCT........................................................................................ | 28 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................GTTCAAGAACGGTGCTCAGAGTTT................................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................TAGTCCCTCGTCCTTTTGAAATCGAGAA......................................... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................GGACGGTGCTCAGAGTTTAACTGTCCT........................................................................................ | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................TTTAAGAACGGTGCTCAGAGTTTA................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 08/21/2014 at 10:12 AM