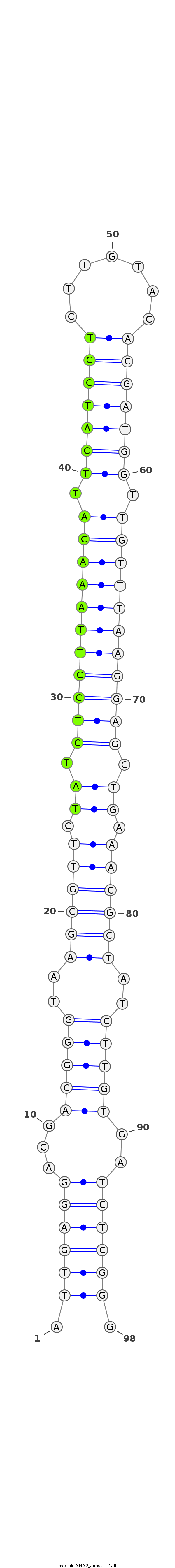
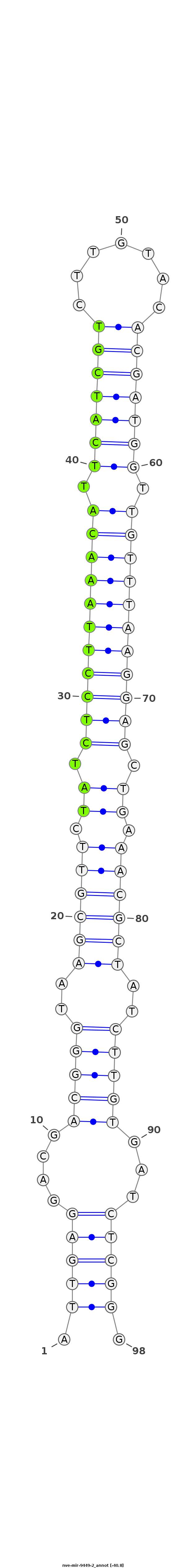
ID:nve-mir-9449-2 |
Coordinate:scaffold_82:62139-62206 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.8 | -40.8 | -40.7 |
 |
 |
 |
| mature | star |
|
CTCTCGTTTTTTACTTGTTGGGAGGGACGTGGACGATTGAGGACGACGGGTAAGCGTTCTATCTCCTTAAACATTCATCGTCTTGTACACGATGGTTGTTTAAGGAGCTGAAACGCTATCTTGTGATCTCGGGGATGACTCGGTGTTCTCCAAGAAGAGGTCTAAGGA
***********************************.((((((...(((((..((((((.((.(((((((((((.(((((((.......))))))).))))))))))).)).))))))..)))))..)))))).*********************************** |
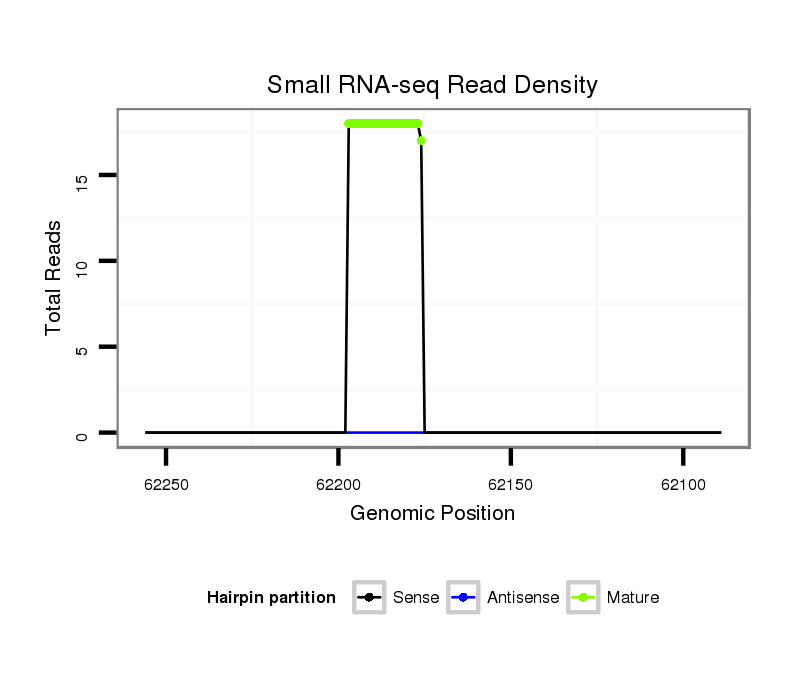
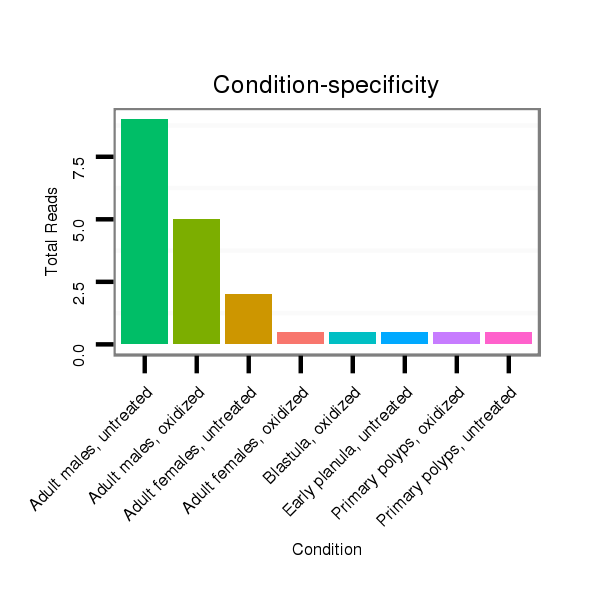
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039754 "Adult males, untreated" |
SRR039755 "Adult males, oxidized" |
SRR039731 "Adult females, untreated" |
SRR039764 "Primary polyps, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039729 "Blastula, oxidized" |
SRR039732 "Adult females, oxidized" |
SRR039758 "Early planula, untreated" |
SRR039765 "Primary polyps, oxidized" |
SRR039756 "Gastrula, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
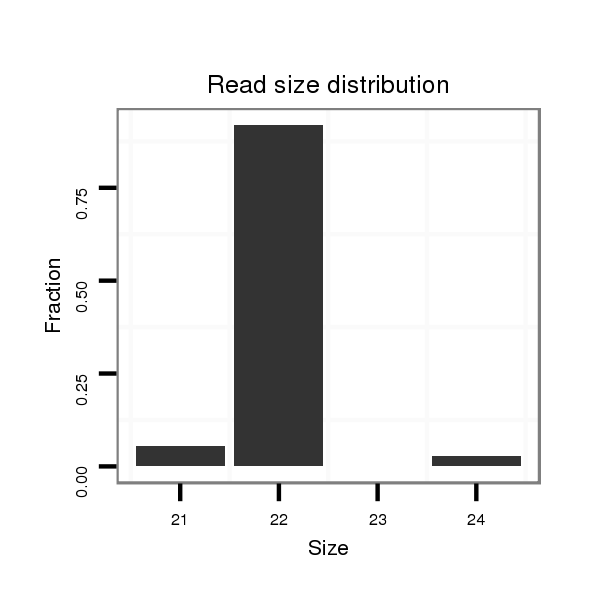
| ...........................................................TATCTCCTTAAACATTCATCGT....................................................................................... | 22 | 0 | 2 | 17.00 | 34 | 17 | 9 | 3 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| ...........................................................TATCTCCTTAAACATTCATCG........................................................................................ | 21 | 0 | 2 | 1.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TATCGCCTTAAAAATTCATCGT....................................................................................... | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TCGTCTTGTACACGATGGAA....................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TATCTCCTTAAACATTCATCGTCT..................................................................................... | 24 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TATATCCTTAAACATGCATCGT....................................................................................... | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TATCTCCTTAAACATTCATCT........................................................................................ | 21 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TATCTTCTTAAACATTCATCGT....................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TATCTCCTTAAACATCCATCGT....................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CTCAAACATCCATCGTCTT.................................................................................... | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CACTAACTTGTGAACTCGG.................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................TGAGCTGAAACGCTATCC............................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
GAGAGCAAAAAATGAACAACCCTCCCTGCACCTGCTAACTCCTGCTGCCCATTCGCAAGATAGAGGAATTTGTAAGTAGCAGAACATGTGCTACCAACAAATTCCTCGACTTTGCGATAGAACACTAGAGCCCCTACTGAGCCACAAGAGGTTCTTCTCCAGATTCCT
***********************************.((((((...(((((..((((((.((.(((((((((((.(((((((.......))))))).))))))))))).)).))))))..)))))..)))))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 08/21/2014 at 10:46 AM