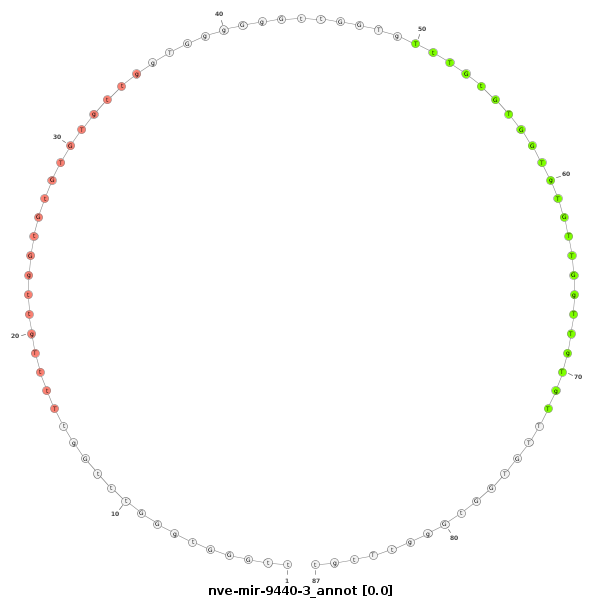
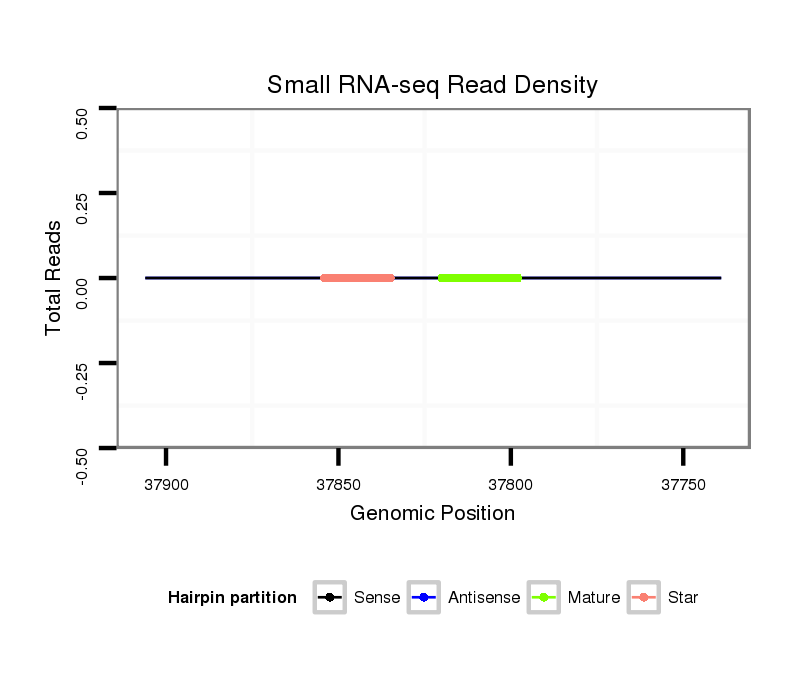
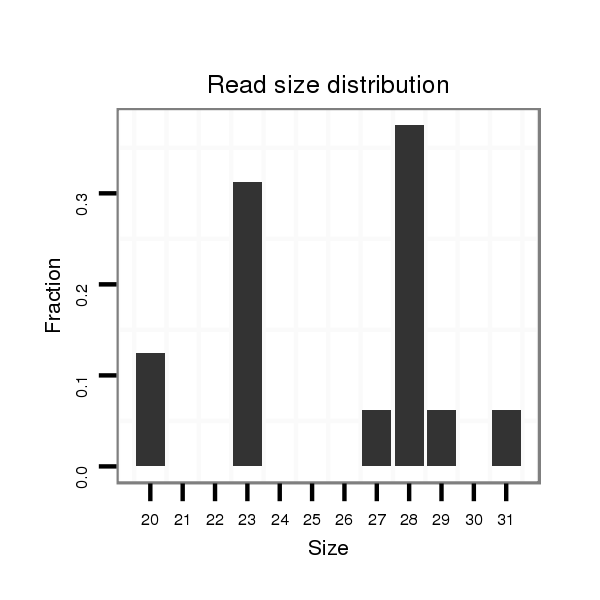
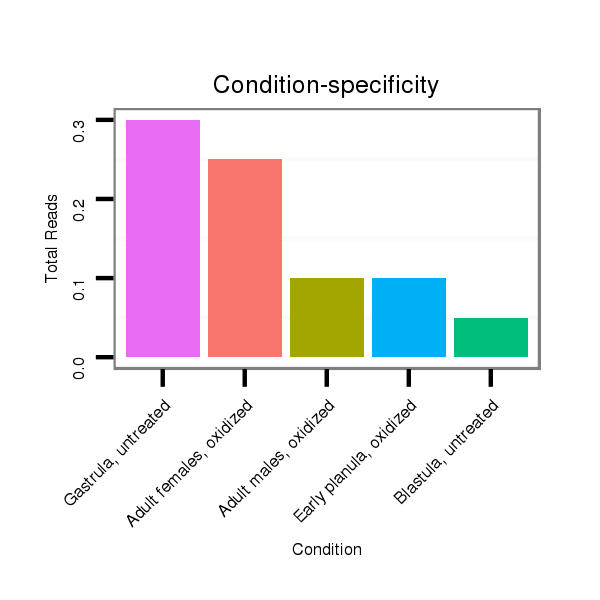
ID:nve-mir-9440-3 |
Coordinate:scaffold_700:37789-37856 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
GtGgttGTGttggTggtgttGtggTTgGTGtTTtGgtttGGGtgGGtttGgtTttTgttgGtGtGTGTgttggTGggGgGttGGTgTtTGtGTGGTgTGTTGgTTgTgTTTGTGGtGggtTtgtgGGgtGGGGgTgGgGtgTgTGtGttGTTgGTGTtggggGtgtTg
*************************************.......................................................................................******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039756 "Gastrula, untreated" |
SRR039761 "Late planula, oxidized" |
SRR039732 "Adult females, oxidized" |
SRR039755 "Adult males, oxidized" |
SRR039759 "Early planula, oxidized" |
SRR039726 "Blastula, untreated" |
SRR039728 "Unfertilized eggs, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................AGGGACGGAAAGCATAATCAACGAGAGTG..................................................................................................... | 29 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TATGAGTGGTCTGTTGCTTCTCT........................................................... | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TATGAGTGGTCTGTTGCTTCTCTTTGTG...................................................... | 28 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................AGGTCTATGAGTCGTCTGTTGCTTTTCT........................................................... | 28 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TAATCAACGAGAGTGTCAAC................................................................................................ | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ............................TGATTAGCAAAGGGACGGA......................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TAATCAACGAGAGTGTCAACCTGCCGCGA....................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TAATCAACGAGAGTGTCAACCTGCCGC......................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................TCAACGAGAGTGTCAACCTGCCGCGAAG..................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................TGAGAAGTTCGTGTACCCCGACATC | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TAATCAACGAGAGTCTCAACCTGCCG.......................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................TAATCAACGAGAGTGTCAACCTGCCGCGAAG..................................................................................... | 31 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
CtCgttCACttggAggtgttCtggAAgCACtAAtCgtttCCCtgCCtttCgtAttAgttgCtCtCACAgttggACggCgCttCCAgAtACtCACCAgACAACgAAgAgAAACACCtCggtAtgtgCCgtCCCCgAgCgCtgAgACtCttCAAgCACAtggggCtgtAg
********************************************.......................................................................................************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039726 "Blastula, untreated" |
SRR039731 "Adult females, untreated" |
SRR039728 "Unfertilized eggs, oxidized" |
SRR039761 "Late planula, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039757 "Gastrula, oxidized" |
SRR039765 "Primary polyps, oxidized" |
SRR039727 "Unfertilized eggs, untreated" |
SRR039729 "Blastula, oxidized" |
SRR039732 "Adult females, oxidized" |
SRR039758 "Early planula, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................CGTTTCCCTGCCTTTCGTATTAGTTGCT.......................................................................................................... | 28 | 0 | 3 | 0.67 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................GCGCTGAGACTCTTCAAGCACATGGGGCT.... | 29 | 0 | 20 | 0.40 | 8 | 0 | 5 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...................................GTTTCCCTGCCTTTCGTATTAGTTGCT.......................................................................................................... | 27 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................CGCTGAGACTCTTCAAGCACATGGGGCT.... | 28 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GGACGGCGCTTCCAGATACTCACCA........................................................................ | 25 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................GACGGCGCTTCCAGATACTCACCA........................................................................ | 24 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................................................CACAGTTGGACGGCGCTTCCAGATACT............................................................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................ATATGTGCCGTCCCCGAGC............................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................CCGAGCGCTGAGACTCTTCAAGCACATGG........ | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TTGGACGGCGCTTCCAGATACTCACCA........................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................GCGCTGAGACTCTTCAGGCACATGGGGCT.... | 29 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CGAGCGCTGAGACTCTTCAAGCATATGG........ | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TGTGCCGTCCCCGAGCGCTGAGACTCT.................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................ATGTGCCGTCCCCGAGCGCTGAGACTCT.................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................CTGAGACTCTTCAAGCACATGGGGCT.... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................ACGGCGCTTCCAGATACTCACCA........................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................GTTGGACGGCGCTTCCAGATACTCACCA........................................................................ | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CGGCGCTTCCAGATACTCACCA........................................................................ | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................CGAGCGCTGACACTCTTCAAGCACATGGGG...... | 30 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GGAGGTGTTCTGGAAGCACTAATCGTTT................................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................AGCGCTGAGACTCTTCAAGCACATGGGGCT.... | 30 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CTTGGAGGTGTTCTGGAAGCACTAATCGT................................................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 08/21/2014 at 10:46 AM