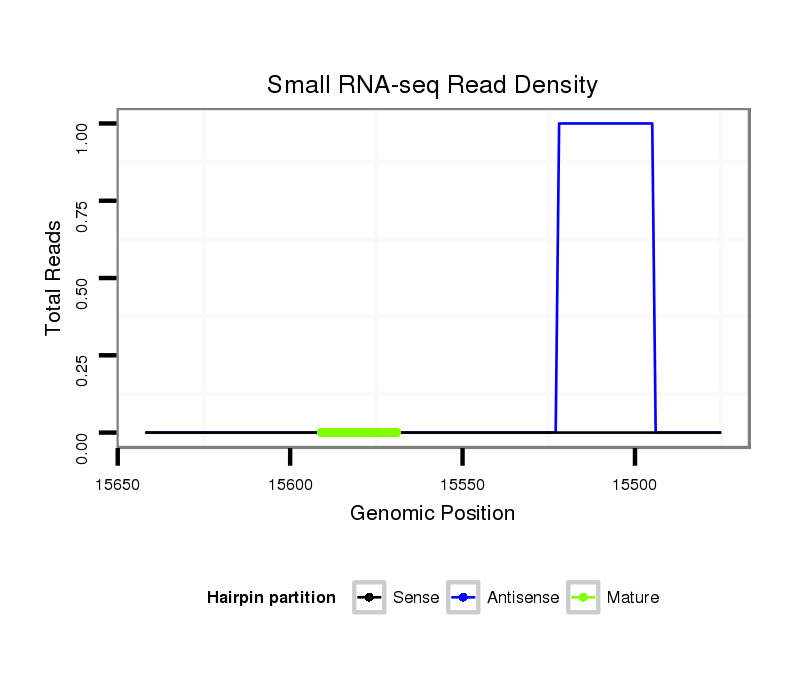
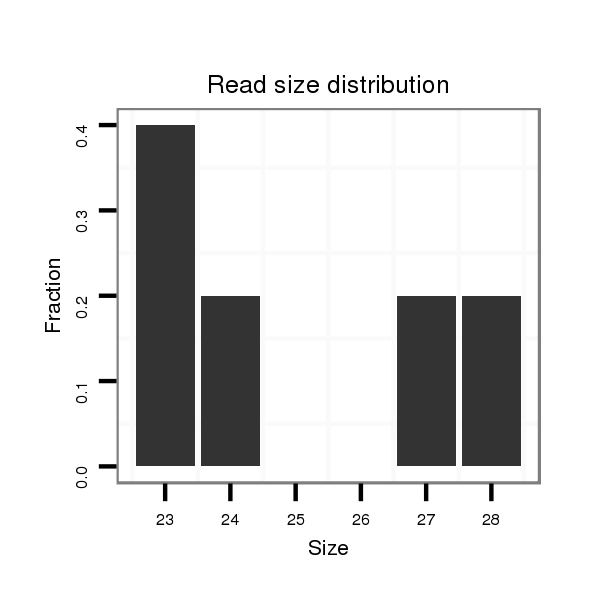
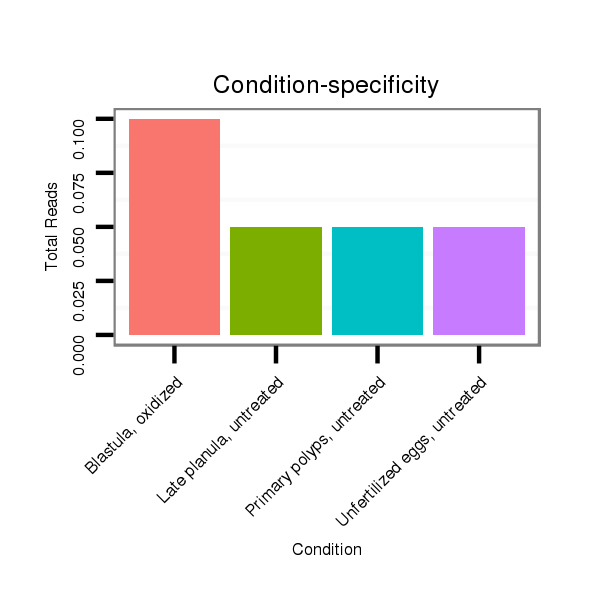
ID:nve-mir-9440-2 |
Coordinate:scaffold_88:15525-15592 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
tGgttGTGttggTggtgttGtggTTgGTGtTTtGgtttGGGtgGGtgtGgtTttTgttgGtGtGTGTgttggTGggGgGttGGTgTtTGtGTGGTgTGTTGgTgtTgTTTGTGTtGggtTtgtgGGgtGGtGgTgGgGtgTgTGtGttGTTgGTGTtggggGtgtTgt
***********************************..................................................................................................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039764 "Primary polyps, untreated" |
SRR039757 "Gastrula, oxidized" |
SRR039727 "Unfertilized eggs, untreated" |
SRR039729 "Blastula, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039731 "Adult females, untreated" |
SRR039760 "Late planula, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................AGGACTATGAGGGGTCTGTTGATCATCT............................................................ | 28 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................TATGAGAGGTCTGTTGCACATCTTTGT........................................................ | 27 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCATAATCAACGAGAGTGTCAACCTGC............................................................................................. | 27 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...........................TGATTAGCAAAGGGACGGA.......................................................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ...................................................TAATCAACGAGAGTGTCAACCTG.............................................................................................. | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...............................................AGCATAATCAACGAGAGTGTCAACCTGC............................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................TAATCAACGAGAGTGTCTACCTG.............................................................................................. | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................AATCAACGAGAGTGTCAACCTGCC............................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................AATCAACGAGAGTGTCAACCTGCCGCG......................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................GTGGTCTGTTGCTCATCTTTGTGG...................................................... | 24 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TAATCAACGAGAGTGTCAACCTGCCGCG......................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
tCgttCACttggAggtgttCtggAAgCACtAAtCgtttCCCtgCCtgtCgtAttAgttgCtCtCACAgttggACggCgCttCCAgAtACtCACCAgACAACgAgtAgAAACACAtCggtAtgtgCCgtCCtCgAgCgCtgAgACtCttCAAgCACAtggggCtgtAgt
***********************************..................................................................................................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039728 "Unfertilized eggs, oxidized" |
SRR039731 "Adult females, untreated" |
SRR039727 "Unfertilized eggs, untreated" |
SRR039760 "Late planula, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039754 "Adult males, untreated" |
SRR039757 "Gastrula, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039759 "Early planula, oxidized" |
SRR039755 "Adult males, oxidized" |
SRR039729 "Blastula, oxidized" |
SRR039765 "Primary polyps, oxidized" |
SRR039758 "Early planula, untreated" |
SRR039764 "Primary polyps, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................TGTGCCGTCCTCGAGCGCTGAGACTCTT.................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCGCTGAGACTCTTCAAGCACATGGGGCT..... | 29 | 0 | 20 | 0.40 | 8 | 0 | 5 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CAGTTGGACGGCGCTTCCAGATACTCACCA......................................................................... | 30 | 0 | 20 | 0.20 | 4 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGTTGGACGGCGCTTCCAGATACTCACCA......................................................................... | 29 | 0 | 20 | 0.20 | 4 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CGCTGAGACTCTTCAAGCACATGGGGCT..... | 28 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................GACGGCGCTTCCAGATACTCACCA......................................................................... | 24 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ...................................................................TTTGGACGGCGCTTCCAGATACTCACCA......................................................................... | 28 | 1 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................CGGCGCTTCCAGATACTCACCA......................................................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................GTTTCCCTGCCTGTCGTATTAGTTGCT........................................................................................................... | 27 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................ACACATCGGGATGTGCATT........................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................AGACTCTTCAAGCACATGGGGCTGTAGT | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CTGAGACTCTTCAAGCACATGGGGCT..... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................CGAGCGCTGACACTCTTCAAGCACATGGGG....... | 30 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................GGACGGCGCTTCCAGATACTCACCA......................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................CGTTTCCCTGCCTGTCGGATTAGTTGCT........................................................................................................... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CGCTGAGACTCTTCAAGCACATCGGGCT..... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................CTTTTCCCTGCCTGTCGTATTAGTTGCT........................................................................................................... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................ACTAATCGTTTCCCTGCCTGTCGTAT................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GCGCTGAGACTCTTCAAGCACACGGGGCT..... | 29 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................ACGGCGCTTCCAGATCCTCACCA......................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................GTTTCCCTGCCTGTCGTA.................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................TGCCTGTCGTATTAGTTGCT........................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGCGCTGAGACTCTTCAAGCACATGGGGCT..... | 30 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TTTCCCTGCCTGTCGTATTAGTTGCT........................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CTTGGAGGTGTTCTGGAAGCACTAATCGT.................................................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TTGGACGGCGCTTCCAGATACTCACCA......................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................GGACGGCGCTTCCAGATACTCAC........................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........GGAGGTGTTCTGGAAGCACTAATCGTTT.................................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................ACAGTTGGACGGCGCTTCCAGATACT.............................................................................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........TCTGAGGTGTTCTGGAAGCACT.......................................................................................................................................... | 22 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 08/21/2014 at 10:52 AM