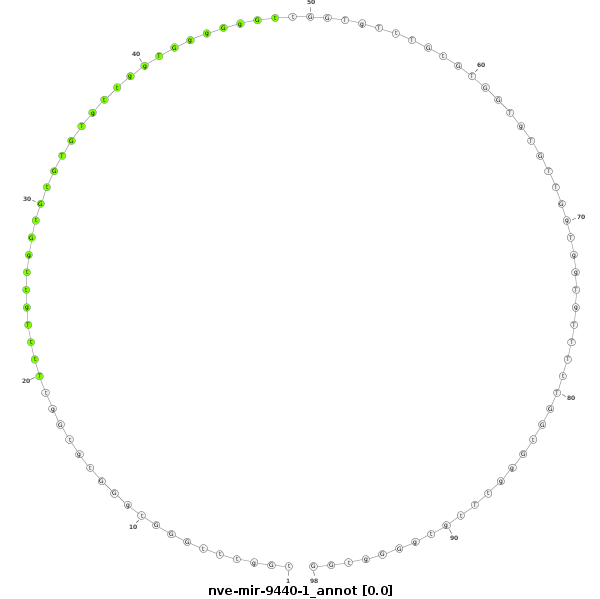
ID:nve-mir-9440-1 |
Coordinate:scaffold_39:1060822-1060889 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
tgGTGgttGTGttggTTgtgttGtggTTgGTGtTTtGgtttGGGtgGGtgtGgtTttTgttgGtGtGTGTgttggTGggGgGttGGTgTtTGtGTGGTgTGTTGgTggTgTTTtTGGtGggtTtgtgGGgtGGGGgTgGgGtgTgTGtGttGTTgGTGTtggggGtgt
***********************************..................................................................................................*********************************** |
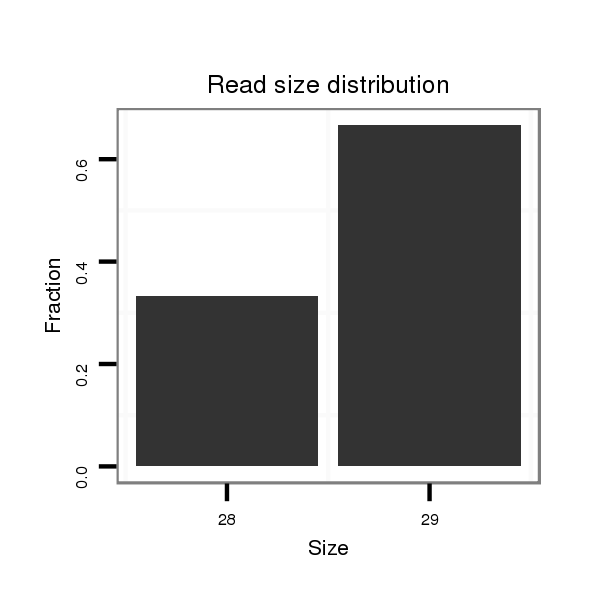
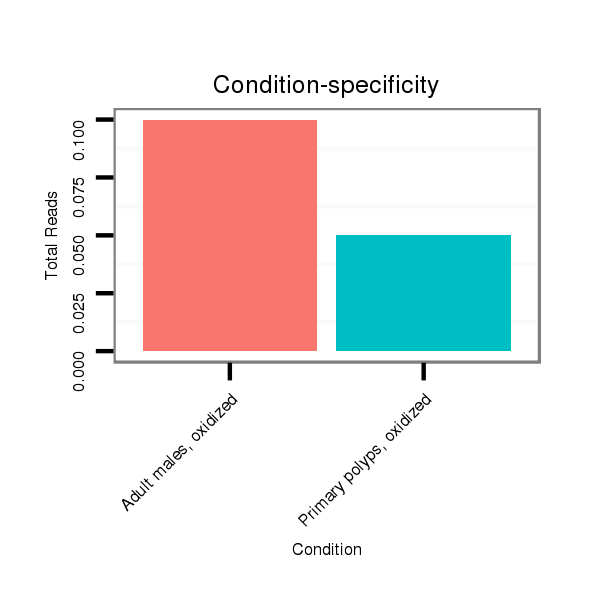
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039755 "Adult males, oxidized" |
SRR039754 "Adult males, untreated" |
SRR039758 "Early planula, untreated" |
SRR039759 "Early planula, oxidized" |
SRR039761 "Late planula, oxidized" |
SRR039765 "Primary polyps, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................TAATCAACGAGAGTGTCAACCTGCCGCGA..................................................................................... | 29 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................TTCGTGATTAGCAAAGGGACGGACAGCA.................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................TTCGTGATTAGCAAAGGGACGGAC...................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............TTCACAAGACCTTCGTGATAT.................................................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........TGAACCTTCACAAGACCTTCGTGATTAGCAA................................................................................................................................ | 31 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................TAATCAACGAGAGTGTCAACCTGCCGCG...................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
tgCACgttCACttggAAgtgttCtggAAgCACtAAtCgtttCCCtgCCtgtCgtAttAgttgCtCtCACAgttggACggCgCttCCAgAtACtCACCAgACAACgAggAgAAAtACCtCggtAtgtgCCgtCCCCgAgCgCtgAgACtCttCAAgCACAtggggCtgt
***********************************..................................................................................................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039731 "Adult females, untreated" |
SRR039757 "Gastrula, oxidized" |
SRR039760 "Late planula, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039727 "Unfertilized eggs, untreated" |
SRR039758 "Early planula, untreated" |
SRR039759 "Early planula, oxidized" |
SRR039754 "Adult males, untreated" |
SRR039755 "Adult males, oxidized" |
SRR039729 "Blastula, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039765 "Primary polyps, oxidized" |
SRR039726 "Blastula, untreated" |
SRR039728 "Unfertilized eggs, oxidized" |
SRR039764 "Primary polyps, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................GCGCTGAGACTCTTCAAGCACATGGGGCT.. | 29 | 0 | 20 | 0.55 | 11 | 5 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................AGTTGGACGGCGCTTCCAGATACTCACCA...................................................................... | 29 | 0 | 20 | 0.35 | 7 | 2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................CGCTGAGACTCTTCAAGCACATGGGGCT.. | 28 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................CAGTTGGACGGCGCTTCCAGATACTCACCA...................................................................... | 30 | 0 | 20 | 0.20 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TTTGGACGGCGCTTCCAGATACTCACCA...................................................................... | 28 | 1 | 20 | 0.15 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................GACGGCGCTTCCAGATACTCACCA...................................................................... | 24 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................CGGCGCTTCCAGATACTCACCA...................................................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................GTTTCCCTGCCTGTCGTATTAGTTGCT........................................................................................................ | 27 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................CCGAGCGCTGAGACTCTTCAAGCACATGG...... | 29 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGTGTTCTGGAAGCACTAAT.................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................ATATGTGCCGTCCCCGAGC............................. | 19 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ATGTGCCGTCCCCGAGCGCTGAGACTCTT................. | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................GTTTCCCTGCCTGTCGTA................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................ACTAATCGTTTCCCTGCCTGTCGTAT................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................GCGCTGAGACTCTTCAAGCACACGGGGCT.. | 29 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................TTGGACGGCGCTTCCAGATACTCACCA...................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TGTGCCGTCCCCGAGCGCTGAGACTCT.................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................CTTTTCCCTGCCTGTCGTATTAGTTGCT........................................................................................................ | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................GTTTCCCTGCCTGTCGTATTAGTTTCT........................................................................................................ | 27 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................AGCGCTGAGACTCTTCAAGCACATGGGGCT.. | 30 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GGACGGCGCTTCCAGATACTCAC........................................................................ | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ATGTGCCGTCCCCGAGCGCTGAGACTCT.................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CGAGCGCTGAGACTCTTCAAGCACATCG...... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................GGACGGCGCTTCCAGATACTCACCA...................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................TGCCTGTCGTATTAGTTGCT........................................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................ACAGTTGGACGGCGCTTCCAGATACT........................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 08/21/2014 at 10:18 AM