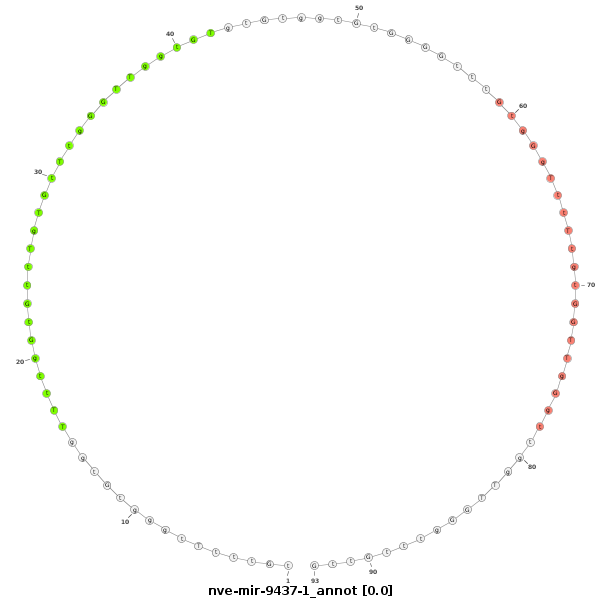
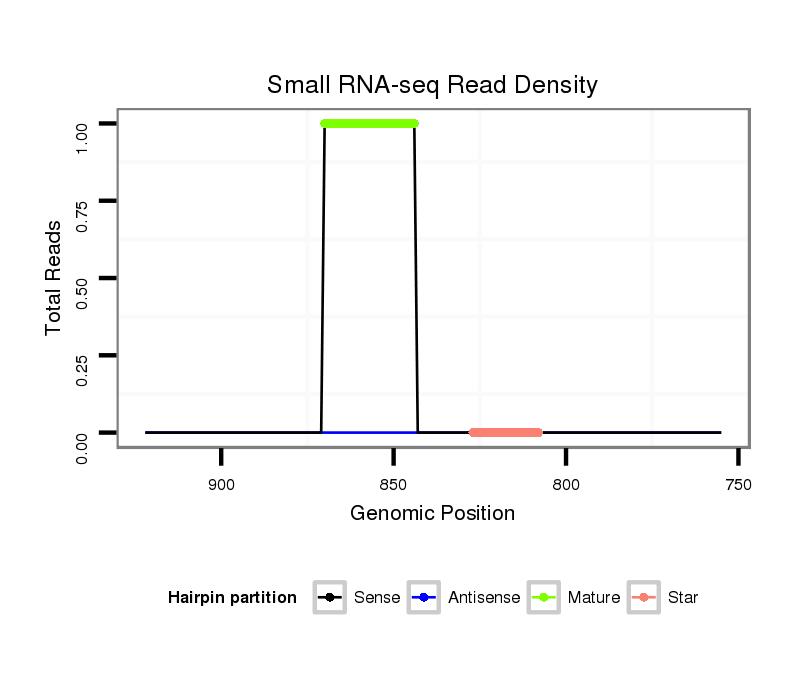
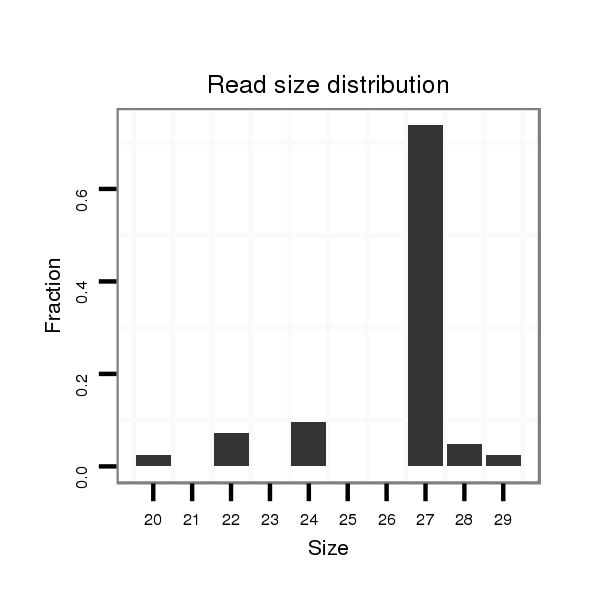
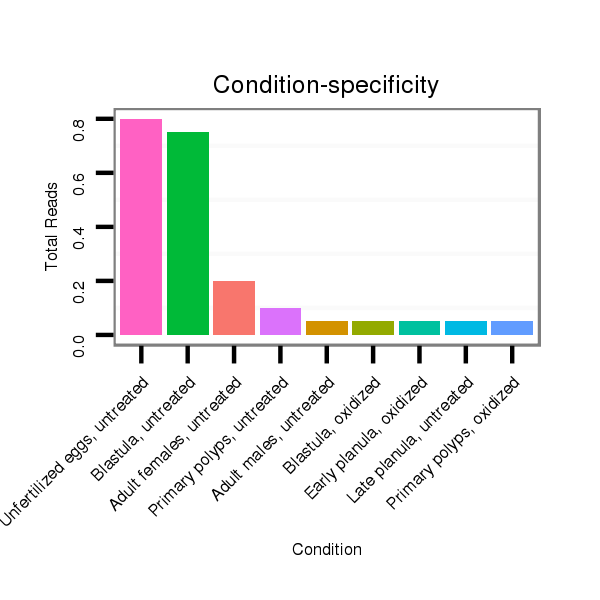
ID:nve-mir-9437-1 |
Coordinate:scaffold_4575:805-872 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
GGTGgtGgGtgttGtgTgGGTgTggtGTgggTgtGGGtGtttTtgggtGtggTTttgGtGttTgTGtTtgGGTTggtGTgtGtggtGtGGGGtttGtgGgTttTtgtGGTTgGgttggTTGGgtttGttGGTTGGtgTtttggttttgtGgTGTTgttGGtGGtgGGG
*************************************.............................................................................................************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039726 "Blastula, untreated" |
SRR039727 "Unfertilized eggs, untreated" |
SRR039731 "Adult females, untreated" |
SRR039759 "Early planula, oxidized" |
SRR039757 "Gastrula, oxidized" |
SRR039760 "Late planula, untreated" |
SRR039754 "Adult males, untreated" |
SRR039761 "Late planula, oxidized" |
SRR039764 "Primary polyps, untreated" |
SRR039728 "Unfertilized eggs, oxidized" |
SRR039729 "Blastula, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039758 "Early planula, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039765 "Primary polyps, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................TTAACGAGAATCTGATACGGTTCCAGT......................................................................................... | 27 | 0 | 20 | 1.55 | 31 | 15 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCGGTCTCCAGTCCCTCAGGGAGAAAT............................................................................................................................. | 27 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTAACGAGAATCTGATACGGTTCC............................................................................................ | 24 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCGGTCTCCAGTCCCTCAGGGAG................................................................................................................................. | 23 | 0 | 20 | 0.15 | 3 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................TTAACGAGAATCTGATACGGTT.............................................................................................. | 22 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCGGTCTCCAGTCCCTCAGGGAGA................................................................................................................................ | 24 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................ACCTTAACGAGAATCTGATACGGTTC............................................................................................. | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCGGTCTCCAGTCCCTCAGGGAGAA............................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................CTTAACGAGAATCTGATACGGTTCCAGT......................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................ACCTTAACGAGAATCTGATACGGTT.............................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTAACGAGAATCTGATACGGTTCCAGTC........................................................................................ | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................CAGACCTTAACGAGAATCTGATACGGTTC............................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................CTTAACGAGAATCTGATACGGTTCCAGTC........................................................................................ | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............AGACTCGGTCTCCAGTCCCTCT...................................................................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................ACCTTAACGAGAATCTGATACGGTTCC............................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCGGTCTCCAGTCCCTCAGGGAGAAATACC.......................................................................................................................... | 30 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTAACGAGAATCTTATACGGTTCCAGTC........................................................................................ | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TCGGTCTCCAGTCCCTCAGGGAGAAATA............................................................................................................................ | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................ACCTTAACGAGAATCTGATACGGT............................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................TCGGTCTCCAGTCCCTCAGGGAGTAATACC.......................................................................................................................... | 30 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTAACGAGAATCAGATACGGTTCCAGTCA....................................................................................... | 29 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TTAACGAGAATCTGATACGGTTCCAGTCT....................................................................................... | 29 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GACGCTAATACAGGTTCGCA..................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CCACgtCgCtgttCtgAgCCAgAggtCAgggAgtCCCtCtttAtgggtCtggAAttgCtCttAgACtAtgCCAAggtCAgtCtggtCtCCCCtttCtgCgAttAtgtCCAAgCgttggAACCgtttCttCCAACCtgAtttggttttgtCgACAAgttCCtCCtgCCC
**************************************.............................................................................................************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039727 "Unfertilized eggs, untreated" |
SRR039726 "Blastula, untreated" |
SRR039729 "Blastula, oxidized" |
SRR039757 "Gastrula, oxidized" |
SRR039756 "Gastrula, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039731 "Adult females, untreated" |
SRR039728 "Unfertilized eggs, oxidized" |
SRR039758 "Early planula, untreated" |
SRR039755 "Adult males, oxidized" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039732 "Adult females, oxidized" |
SRR039754 "Adult males, untreated" |
SRR039764 "Primary polyps, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................CCCTCTTTATGGGTCTGGAATTGCTCTT.......................................................................................................... | 28 | 0 | 20 | 0.60 | 12 | 3 | 0 | 3 | 0 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TCCAACCTGATTTGGTTTTGTCGACAAGTT.......... | 30 | 0 | 6 | 0.33 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................CTTTATGGGTCTGGAATTGCTCTT.......................................................................................................... | 24 | 0 | 20 | 0.25 | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..ACGTCGCTGTTCTGAGCCAGAGGT.............................................................................................................................................. | 24 | 0 | 20 | 0.20 | 4 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TCAGTCTGGTCTCCCCTTTCTGCGATTAT............................................................... | 29 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................CAGTCTGGTCTCCCCTTTCTGCGATTAT............................................................... | 28 | 0 | 20 | 0.20 | 4 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................AGTCTGGTCTCCCCTTTCTGCGATTAT............................................................... | 27 | 0 | 20 | 0.15 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...CGTCGCTGTTCTGAGCCAGAGGT.............................................................................................................................................. | 23 | 0 | 20 | 0.15 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................ATGGGTCTGGAATTGCTCTTAGACTAT................................................................................................... | 27 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....GTCGCTGTTCTGAGCCAGAGGT.............................................................................................................................................. | 22 | 0 | 20 | 0.10 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TGGTCTCCCCTTTCTGCGATTAT............................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................CCTCTTTATGGGTCTGGAATTGCTCTT.......................................................................................................... | 27 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................TGGGTCTGGAATTGCTCTTAGACTAT................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................CTCTTTATGGGTCTGGAATTGCTCTT.......................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................TTTATGGGTCTGGAATTGCTCTT.......................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CGACCTGATTTGGTTTTGTCGACAAGTT.......... | 28 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................CTTTATGGGTCTGGAATTGCTCTTAGA....................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................TCCCTCTTTATGGTTCTGGAATTGCTCTT.......................................................................................................... | 29 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................CTGGTCTCCCCTTTCTGCGATTAT............................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGGGTCTGGAATGGCTCTT.......................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................TTGCTCTTAGACTATGCCAAGGTCAGTCT..................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................AGTCTGGTCTCCCCTTGCTGCGATTAT............................................................... | 27 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .CACGTCGCTGTTCTGAGCCAGAGG............................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................ATGGGTCTGGAATTGCTCTTAGACT..................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................TCTGGTCTCCCCTTTCTGCGATTAT............................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CTGGAATTGCTCTTAGACTATGCCAAGGTC.......................................................................................... | 30 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................TCCCTCTTTATGGGTCTGGAATTGCTCTT.......................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................CTTTATGGGTCTGGAATTGCTCTTAGACT..................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 08/21/2014 at 10:27 AM