
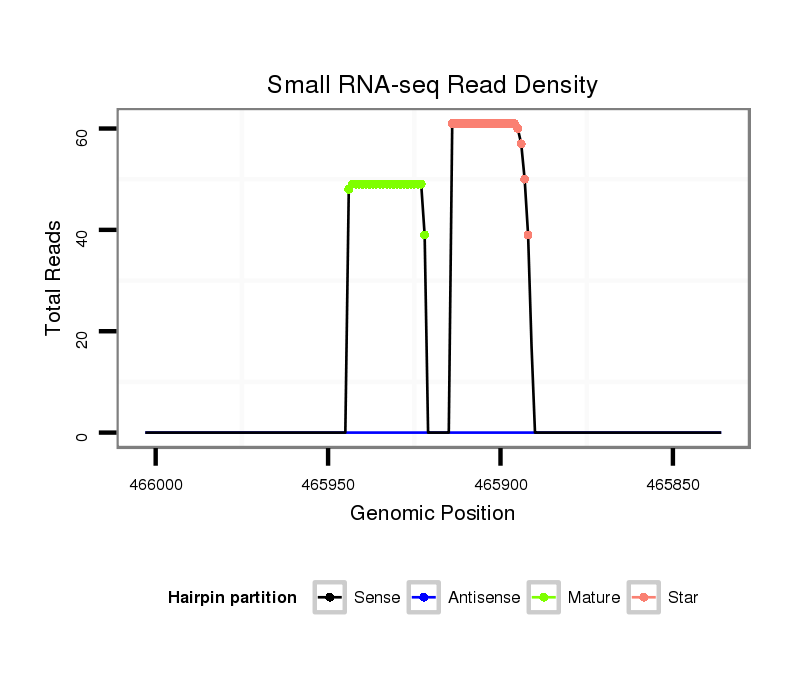
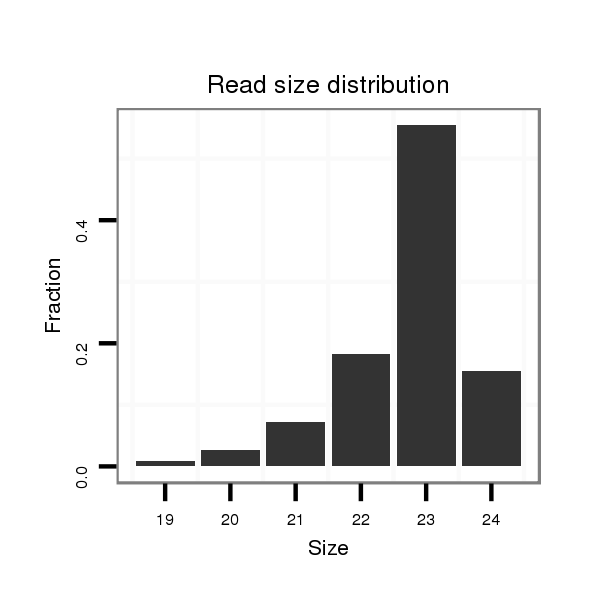
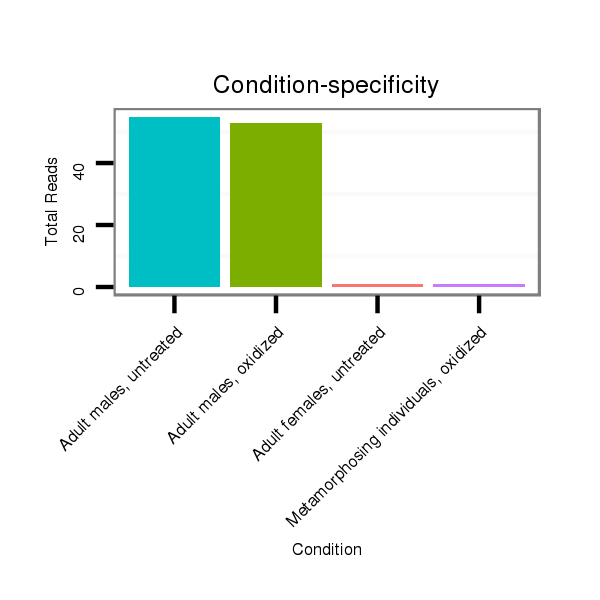
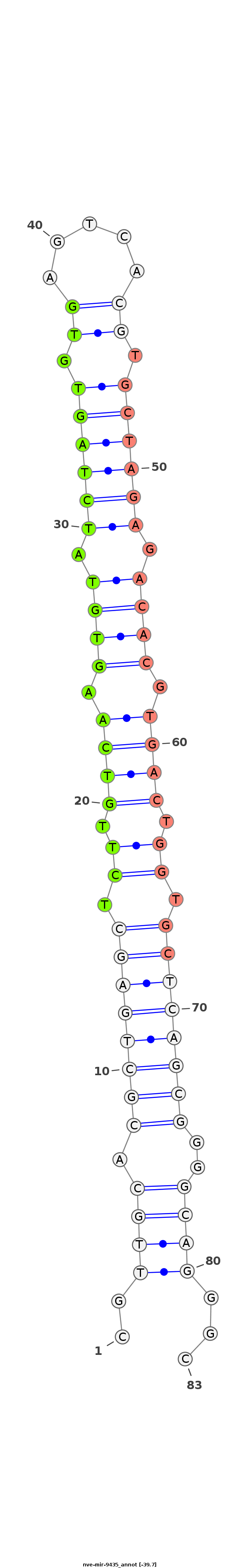
ID:nve-mir-9435 |
Coordinate:scaffold_37:465886-465953 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.0 | -39.7 | -39.7 |
 |
 |
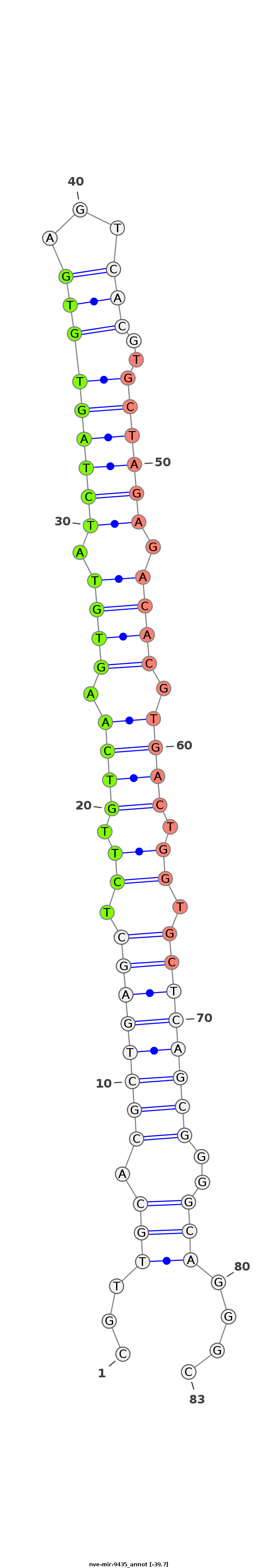 |
|
GACGACACGCAAAGGAGTGGTGCCTACATCCGGGATACTTACATCGTTGCACGCTGAGCTCTTGTCAAGTGTATCTAGTGTGAGTCACGTGCTAGAGACACGTGACTGGTGCTCAGCGGGGCAGGGCGAGTCGGTATGAGGTTAGTAGTTGTGTCGTGAGTTTAAGGT
********************************************..((((.((((((((.((.((((.((((.(((((((((...)))..)))))).)))).)))).)).))))))))..))))...***************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039754 "Adult males, untreated" |
SRR039755 "Adult males, oxidized" |
SRR039726 "Blastula, untreated" |
SRR039729 "Blastula, oxidized" |
SRR039731 "Adult females, untreated" |
SRR039763 "Metamorphosing individuals, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TCTTGTCAAGTGTATCTAGTGTG...................................................................................... | 23 | 0 | 1 | 39.00 | 39 | 26 | 12 | 0 | 0 | 0 | 1 |
| .........................................................................................TGCTAGAGACACGTGACTGGTGC........................................................ | 23 | 0 | 1 | 22.00 | 22 | 6 | 15 | 0 | 0 | 1 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGGTGCT....................................................... | 24 | 0 | 1 | 17.00 | 17 | 8 | 9 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGGTG......................................................... | 22 | 0 | 1 | 11.00 | 11 | 4 | 7 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGTCAAGTGTATCTAGTGT....................................................................................... | 22 | 0 | 1 | 9.00 | 9 | 7 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGGT.......................................................... | 21 | 0 | 1 | 7.00 | 7 | 1 | 6 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGTCAAGTGTATCTAGTGTGT..................................................................................... | 24 | 1 | 1 | 5.00 | 5 | 2 | 3 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGGTGCTT...................................................... | 25 | 1 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGG........................................................... | 20 | 0 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGTCAAGTGTATCTAGTTTG...................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TTCTAGAGACACGTGACTGGTGCT....................................................... | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGGTGT........................................................ | 23 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGTCCAGTGTATCTAGTGT....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................TCTTGTCGAGGGTATCTAGTGTG...................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACGGGTGC........................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................TCTGGTCAAGTGTAGCTAGTGTG...................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TAATAGAGACACGTGACTGGT.......................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTG............................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TTCTAGAGACACGTGACTTGTTC........................................................ | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGGTTT........................................................ | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGCGACTGGCGCC....................................................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGTCAAGGGTATCTAGTGTG...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGGCTGGTGC........................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGTCAAGTTTGTCTAGTGTG...................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTGGTGTA....................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGCCAAGTGTATCTAGTGT....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGTGACTTGCGCTT...................................................... | 25 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................CCTTGTCAAGTGTATCTAGTGTG...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGATACGAGACTGGTG......................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................CTTGTCAAGTGTATCTAGTGT....................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TCTTGTCAAGTGTATCTAGTGTTT..................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TGCTAGAGACACGCGACTGGTTC........................................................ | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TGTTAGAGACACGTGACTGGTGC........................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGATAGAGACACGTGACTGGT.......................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CTGCTGTGCGTTTCCTCACCACGGATGTAGGCCCTATGAATGTAGCAACGTGCGACTCGAGAACAGTTCACATAGATCACACTCAGTGCACGATCTCTGTGCACTGACCACGAGTCGCCCCGTCCCGCTCAGCCATACTCCAATCATCAACACAGCACTCAAATTCCA
*****************************************..((((.((((((((.((.((((.((((.(((((((((...)))..)))))).)))).)))).)).))))))))..))))...******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039731 "Adult females, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................GCCGCGGCCCGCTCAGCCCTAC.............................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................ACCACGGATTTGGGCCCT..................................................................................................................................... | 18 | 2 | 4 | 0.50 | 2 | 0 | 1 | 1 |
| .......................GATGTATGCCCTAAGAGTG.............................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
Generated: 08/21/2014 at 10:06 AM