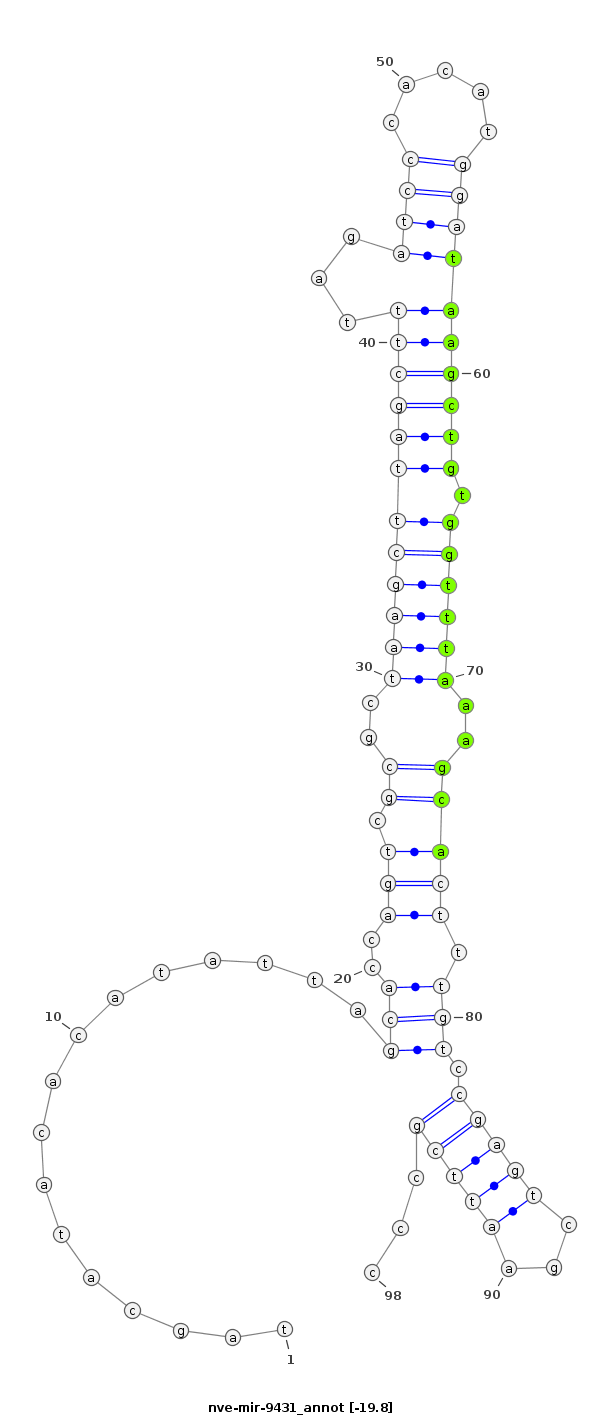

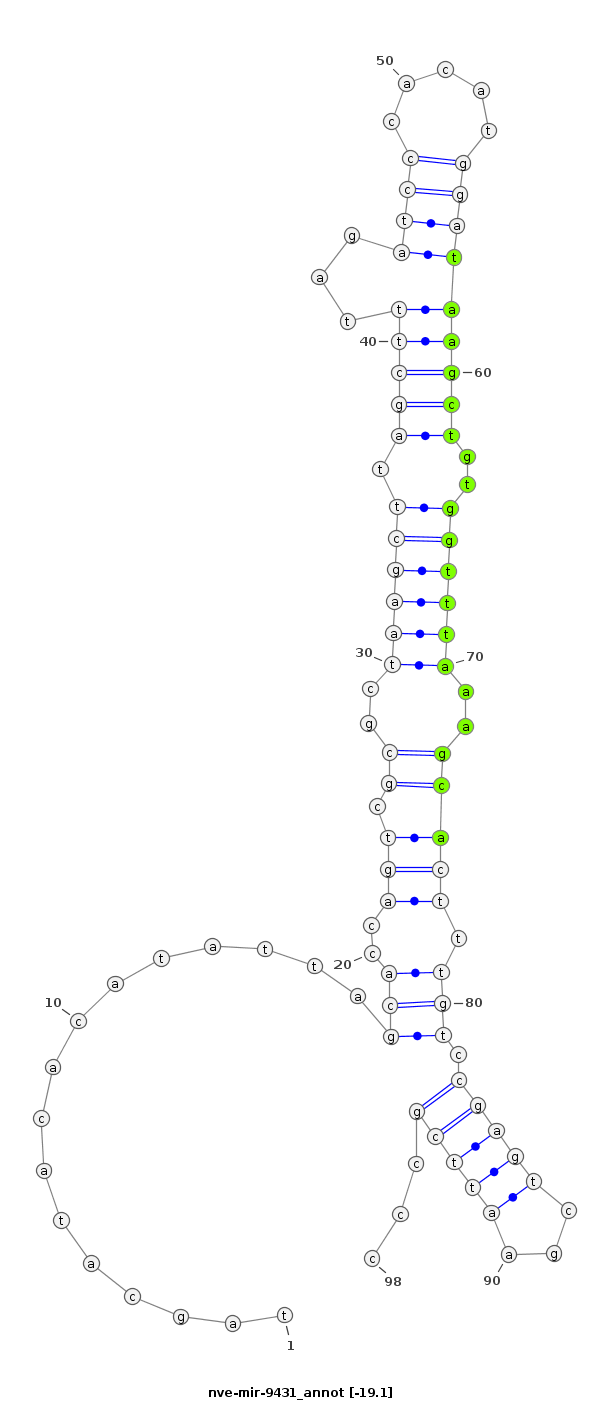
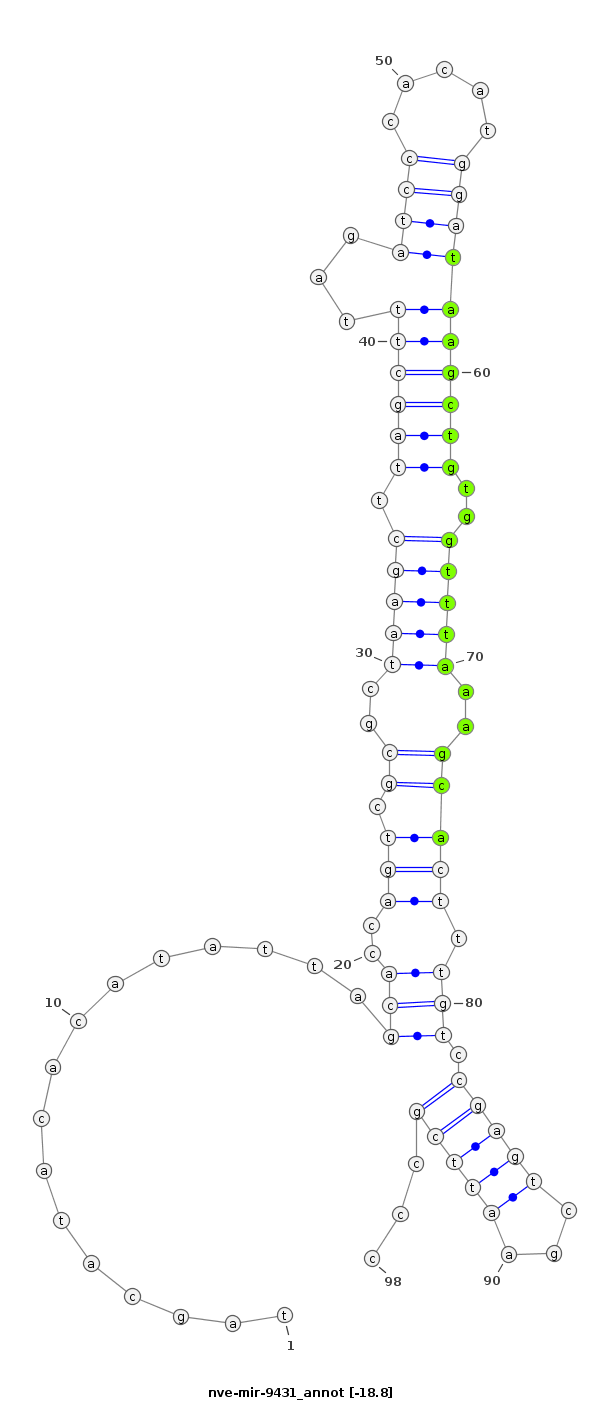
ID:nve-mir-9431 |
Coordinate:scaffold_106:684146-684213 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -19.1 | -18.8 | -17.8 |
 |
 |
 |
|
ACATTGGACGAAAAAATACGGAATACGGAgcacactagcatacacatattagcaccagtcgcgctaagcttagctttagatcccacatggataagctgtggtttaaagcactttgtccgagtcgaattcgccctgcagtttattgccgataaagtttatctaaggcaa
***********************************................(((..(((.((..((((((((((((...((((.....)))))))))).))))))..))))).))).(((((...)))))...*********************************** |
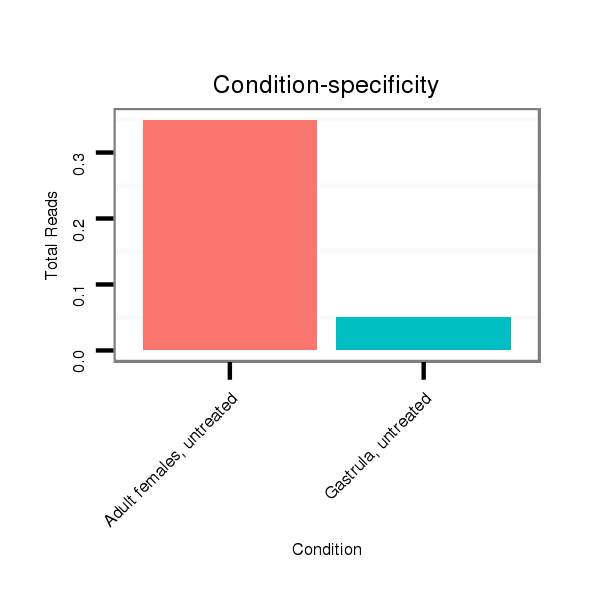
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039731 "Adult females, untreated" |
SRR039754 "Adult males, untreated" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039761 "Late planula, oxidized" |
SRR039756 "Gastrula, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................TAAGCTGTGGTTTAAAGCA.......................................................... | 19 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 |
| ..........................................................................TCTAGATCCCACATGGATAA.......................................................................... | 20 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TAAGCTGTGGTTTAAAGCT.......................................................... | 19 | 1 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................TAAGCTGTGGTTTAAAGCACTAT...................................................... | 23 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAAGCTGTGGTTTAAAGCACAAT...................................................... | 23 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................TCTAGGTCCCACATGGATAA.......................................................................... | 20 | 2 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................TTAGATCTCACATGAATAAAC........................................................................ | 21 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................TAAGCTGTGGTTTAAAGCAC......................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................AGCTCTAGATCCCACATGGATA........................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TGTAACCTGCTTTTTTATGCCTTATGCCTgGTGTGtTgGTtTGTGTtTttTgGTGGTgtGgGgGtTTgGttTgGtttTgTtGGGTGTtggTtTTgGtgtggtttTTTgGTGtttgtGGgTgtGgTTttGgGGGtgGTgtttTttgGGgTtTTTgtttTtGtTTggGTT
***********************************................(((..(((.((..((((((((((((...((((.....)))))))))).))))))..))))).))).(((((...)))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039754 "Adult males, untreated" |
SRR039758 "Early planula, untreated" |
SRR039756 "Gastrula, untreated" |
SRR039759 "Early planula, oxidized" |
SRR039764 "Primary polyps, untreated" |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................AGCTTAAGCGGGACGTCAA............................ | 19 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................ATTCTCAGCTTAAGCGGGCCG................................ | 21 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................AATCGAGATCTAGGGTGTACCT............................................................................. | 22 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................AATTTCGTGAAACACGCTCAGCTT.......................................... | 24 | 1 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................ACACCAAATTTCGTGAAAC..................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................AGCGGGGCGTCAAATAAGGC..................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................TTCGACACCAAATTTCGTGTA....................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 08/21/2014 at 09:07 AM