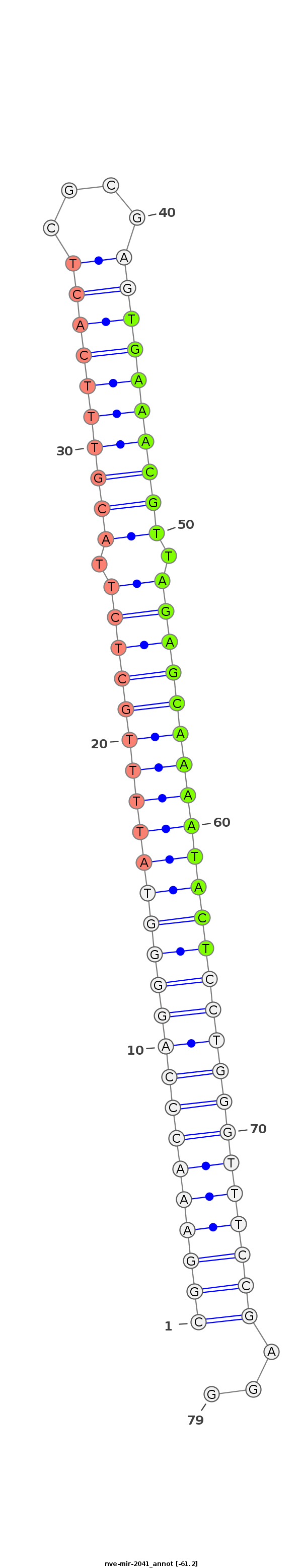

ID:nve-mir-2041 |
Coordinate:scaffold_19:418369-418456 - |
Confidence:Known |
Type:Unknown |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
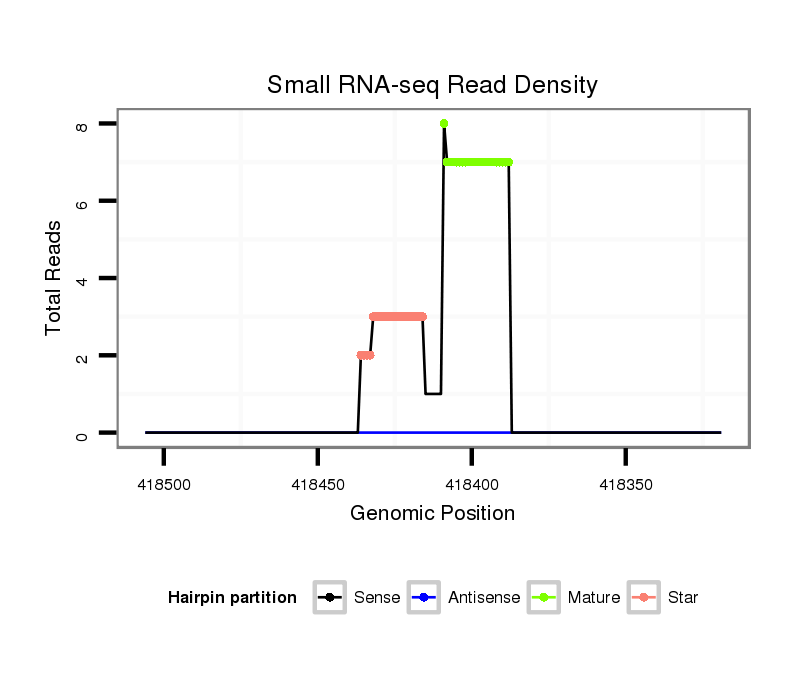
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| mature | star |
|
TGTggTtgTgggggGgGGGTTGgGTGTTGTTGTTTTGggttgTgGtTtGCATCCTCGGAAACCCAGGGGTATTTTGCTCTTACGTTTCACTCGCGAGTGAAACGTTAGAGCAAAATACTCCTGGGTTTCCGAGGATGtgTgGtTtGGttTGtgGGtGtGtTgtTGTTtttTttgTgtGGtGttttttt
*******************************************************(((((((((((((((((((((((((.((((((((((....)))))))))).)))))))))))))))))))))))))...****************************************************** |
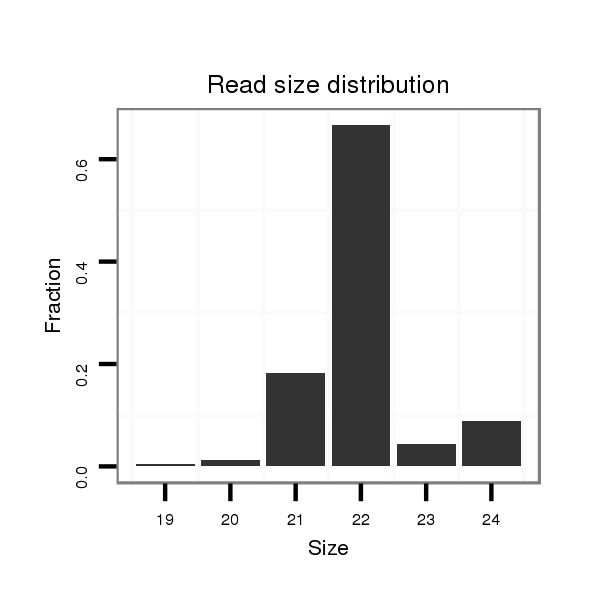
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039731 "Adult females, untreated" |
SRR039754 "Adult males, untreated" |
SRR039764 "Primary polyps, untreated" |
SRR039732 "Adult females, oxidized" |
SRR039765 "Primary polyps, oxidized" |
SRR039762 "Metamorphosing individuals, untreated" |
SRR039755 "Adult males, oxidized" |
SRR039726 "Blastula, untreated" |
SRR039728 "Unfertilized eggs, oxidized" |
SRR039758 "Early planula, untreated" |
SRR039761 "Late planula, oxidized" |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................TGAAACGTTAGAGCAAAATACT..................................................................... | 22 | 0 | 2 | 7.50 | 15 | 7 | 5 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................ATTTTGCTCTTACGTTTCACT................................................................................................. | 21 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................ATTTTTGCTCTTACGTTTCACT................................................................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GCTCTTACGTTTCACTCGCA............................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TTTTTGCTCTTACGTTTCACT................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TGCTCTTACGTTTCACTCGCGAGT.......................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTAGTGCAAAATATT..................................................................... | 22 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................AAACGTTAGAGCAAAATACTCCT.................................................................. | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TGCTCTTACGTTTCACTCGCGATTAT........................................................................................ | 26 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CAAAATACTCCTGGGTTTCT.......................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................AAACGTTAGAGCAAAATACTCT................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTAGAGCAAAATAT...................................................................... | 21 | 1 | 20 | 0.30 | 6 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTAGAGCAAAATAAT..................................................................... | 22 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTAGAGCAAAATACTA.................................................................... | 23 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAAAGTTAGAGTAAAATACT..................................................................... | 22 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................AAACGTTAGAGCAAAATACTTT................................................................... | 22 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTAGAGCAAAATA....................................................................... | 20 | 0 | 20 | 0.15 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................TGAAACGTTAGAGCAAAATTT...................................................................... | 21 | 2 | 20 | 0.15 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTAGAGCAATTT........................................................................ | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACCTTAGAGCAAAATTT...................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTTCAGCAAAATAC...................................................................... | 21 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTTAGAGCAAAATAC...................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TTAAACGTTAGAGCAAAATAC...................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................AAACGTTAGAGCAAAATT....................................................................... | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................CGAGTGAAACGTTAGAGCA............................................................................ | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TGAAACGTGAGAGCAAATTT....................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................GGAATGACGGAGTGATG.......................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
ACAggAtgAgggggCgCCCAACgCACAACAACAAAACggttgAgCtAtCGTAGGAGCCTTTGGGTCCCCATAAAACGAGAATGCAAAGTGAGCGCTCACTTTGCAATCTCGTTTTATGAGGACCCAAAGGCTCCTACtgAgCtAtCCttACtgCCtCtCtAgtACAAtttAttgAgtCCtCttttttt
******************************************************(((((((((((((((((((((((((.((((((((((....)))))))))).)))))))))))))))))))))))))...******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR039755 "Adult males, oxidized" |
SRR039763 "Metamorphosing individuals, oxidized" |
SRR039764 "Primary polyps, untreated" |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................TGAGTCCTCTTTTTT. | 15 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ..........................................AGCTATCCTAGCAGCCT................................................................................................................................. | 17 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 |
| .......................................................GCCTTGGGGTCCCCATAAAAC................................................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 |
Generated: 08/21/2014 at 09:34 AM