
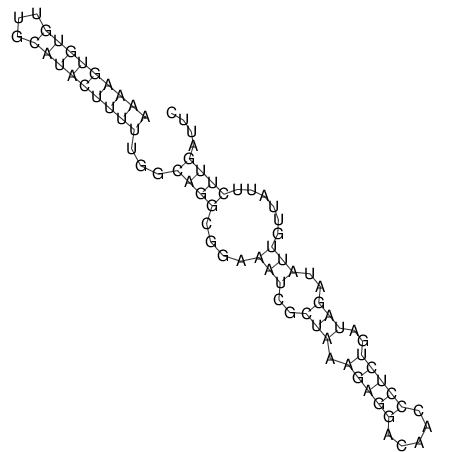
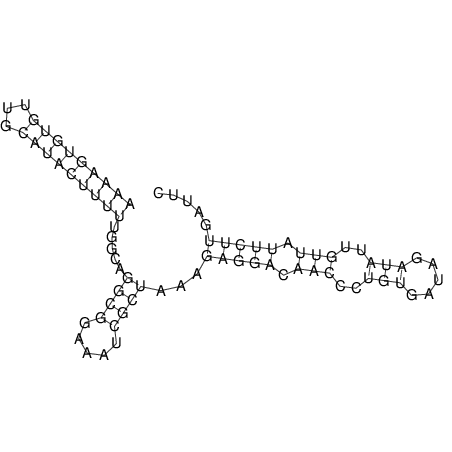



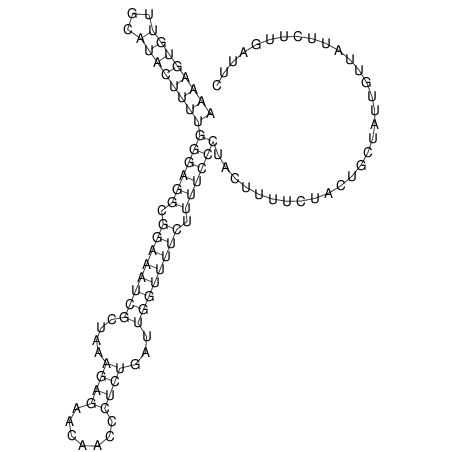
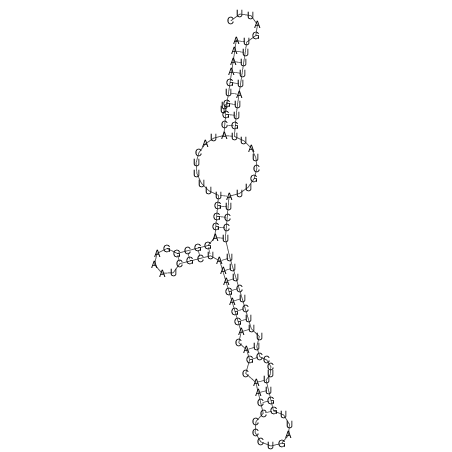
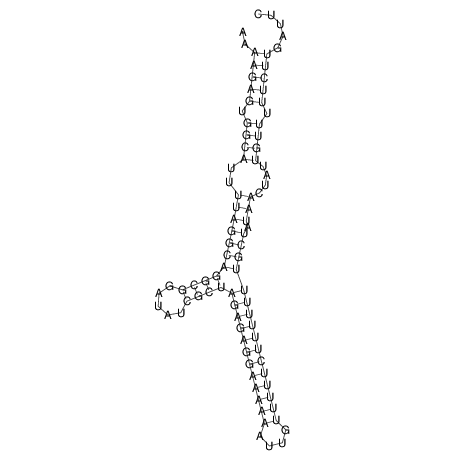


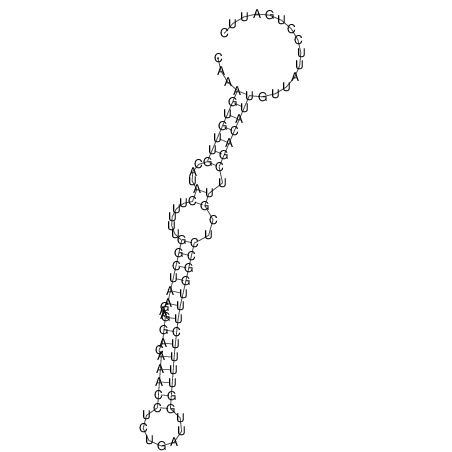
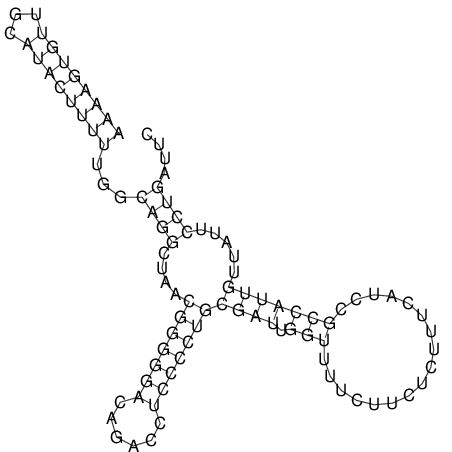
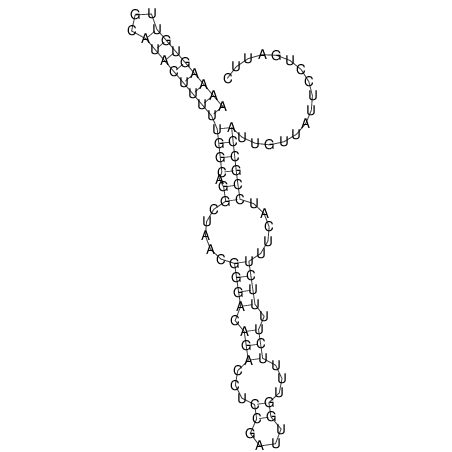
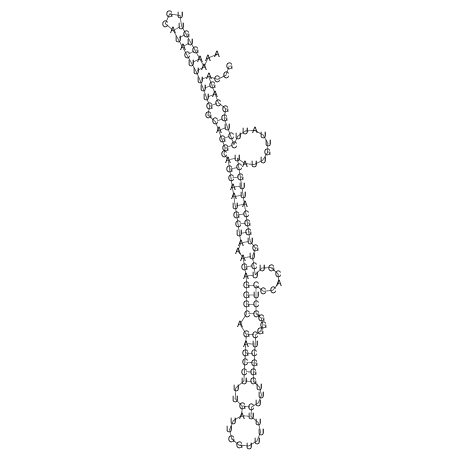
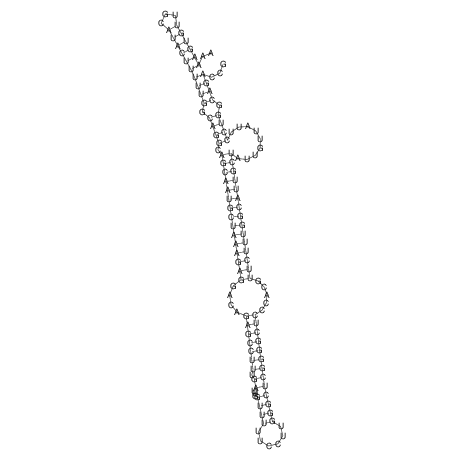
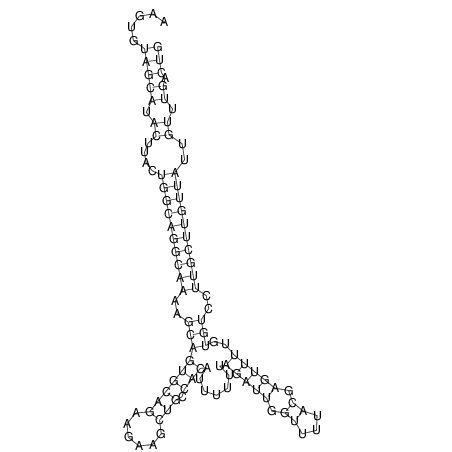
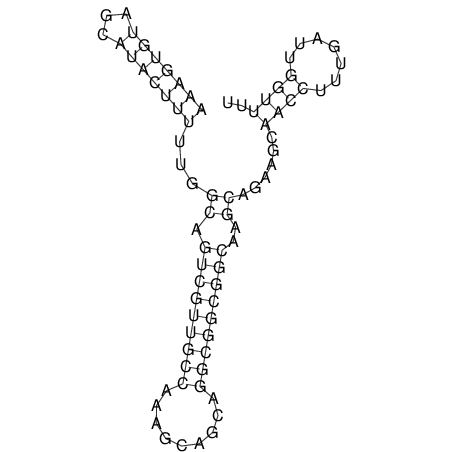
View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
sblock26534 |
chr2L:12284769-12284877 + |
Candidate |
|---|---|---|
| [View on UCSC Genome Browser {Cornell Mirror}] | intron |
Legend:
mature strand at position 12284832-12284853| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr2L:12284754-12284892 + | 1 | AAA----------AGTTCCTTGCGC-CAAAGTGTGTTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCTGATTGGTTTTCTT-TTTGAT-------------T---------TTCGTTCGCCATAG--GTATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TTGA------------------------------CT-GCGTTCTA | |
| droSim1 | chr2L:12084763-12084872 + | LASTZ | 4 | AAA----------AGTTCCTTGCGC-AAAAGTGTGTTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCTGAT----------------------------------------------------AG--ATATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TTGA------------------------------CT-GCGTTGTA |
| droSec1 | super_16:474625-474734 + | LASTZ | 5 | AAA----------AGTTCCTTGCGC-AAAAGTGTGTTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTGTGAT----------------------------------------------------AG--ATATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TGGA------------------------------CT-GCGGTGTA |
| droYak2 | chr2L:8714891-8715022 + | LASTZ | 3 | AAA----------AGTTCTTTTCGC-AAAAGTG--TTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCAGATTGGTTTACTT-TTC----------------T-----CCTTTG------CCATAG--GTTTTGTTATTCTC---------GATTCT-------------------------------------------------------------------------TCGA------------------------------CT-GCCTTGTA |
| droEre2 | scaffold_4929:13133063-13133194 - | LASTZ | 3 | AAC----------AGTTCTTTTCGC-AAAAGTG--TTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGAACAA-------CCCTCAGATTGGTTTTCTT-TTG----------------C-----CCCTTG------CCATAG--GTATAGTTATTCTC---------GATTCC-------------------------------------------------------------------------TTGA------------------------------CT-GCGTTGTA |
| droEug1 | scf7180000409122:667028-667164 + | LASTZ | 4 | AAA----------GTTTTCTTTTGC-GAAAGTG--TTGCATACTTTTCGGCTGGCGGAAA------------------TCGCTA-AAGAGAACAA--C----CCCTCTGATTGGTTTTCTT-TCT----------------T-TGTTCTTTTG------CCATTG--CTATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TAGA------------------------------CT-GGTTTTTT |
| droBia1 | scf7180000302422:707901-708031 - | LASTZ | 4 | AAA----------AGTTG--TTCGC-AAAAGTG--TTGCATACTTT-TGGGAGGCGGAAA------------------TCGCTA-AAGAGAACAA-------CCCTCTGATTGGTTTTCTT-TTC----------------CCT---ACTTTT------CTACTG--CTATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TAGA------------------------------CT-GGGCTGTA |
| droTak1 | scf7180000415865:331344-331479 - | LASTZ | 4 | CAA----------AGTTTTTTCCAC-AAAAGTG--TTGCATACTTTTTGGGAGGCGGAAA------------------TCGCTA-AAGAGGACAG--CAA--CCCCCTGATTGGTTTCCCTTTTC----------------T-----CTTTTC------CTATTG--CTATTGTTATTTTT---------GATTCT-------------------------------------------------------------------------TCAA------------------------------CT-GGGCTGTA |
| droEle1 | scf7180000491028:2849763-2849871 - | LASTZ | 4 | AAA----------AGTTTC-ATTGC-AAAAGAG--TGGCAT---TTTAGGCAGGCGGATA------------------TCGCTA-GAGAGGAAAA-------A------ATTGTTTTTCTT-TT-------------------------TTTG------CTATAA--CTATTGTTTTTCTT---------GATTCT-------------------------------------------------------------------------TATA------------------------------T---------- |
| droRho1 | scf7180000780043:108180-108289 + | LASTZ | 4 | AAA----------AGTTGT-TTCGC-AAAAGTG--TTGCATATTTTTTGGCAGGCGGATA------------------TCGATA-AAGAGGACAA-------CCTTCTT---------C---TTC----------------T-----CTTTTG------CTATTG--CTAATGATATTCT--------------------------------------------------------------------------------------------A------------------------------CT-GTAATTTA |
| droFic1 | scf7180000454015:137236-137368 + | LASTZ | 4 | AAA----------GTTTTCCTTCGC-AAAAGTG--TTGCATACTT-TTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCTGATTGGTTTTCTT-TTT----------------C-----TTTTTG------CTGTTCCTCTATTCTTATTTTC---------GATTCG-------------------------------------------------------------------------TCGT------------------------------CT-GTGGTGTA |
| droKik1 | scf7180000301476:370791-370905 + | LASTZ | 4 | CGT----------TTTACCACCCAC-CAAAGTG--TTGCATACTTTTTGG-------------------------------CTA-AGAAGGACAA-------ACCTCTGATTGGTTTTCTT-TGG----------------CCT---CGT------------TCG--ACATTGTTATTCCT---------GATTCT-------------------------------------------------------------------------TCGA------------------------------CT-GGGGTGTA |
| droAna3 | scaffold_12916:13110146-13110270 - | LASTZ | 5 | ACC----------CGTTTG-CCTGC-AAAAGTG--TTGCATACTTTTTGGCAGGCTAAC-------------------G--------GGGGACAG--ACCTCCCCTGCGATTGGTTTTCTT-CTC----------------T---------TT------CATCCG--CCATTGTTATTCCT---------GATTCG-------------------------------------------------------------------------GAGA------------------------------GTCGCATTCGA |
| droBip1 | scf7180000396580:411008-411118 + | LASTZ | 4 | ACC----------CGTTTT-CCGGC-AAAAGTG--TTGCATACTTTTTGGCAGGC---------------------------TA-ACG-GGACAG-------ACCTCCGATTGGTTTTCTT-TTC----------------T---------TT------CATCCG--CCATTGTTATTCCT---------GATTCG-------------------------------------------------------------------------GGGA------------------------------AT-------CA |
| dp4 | chr4_group3:5836352-5836516 - | LASTZ | 1 | GAG----------TGT------CGC-AAAAGTG--TTGCATACTTTTTGGCAGGCAGCAA------------------T-GCTA-AAGAGGGCAG--A----GCCTTTGATTGGTTTTTCT-TTGGGCTCGGGGCTCCCAC---GT-TCTGTG------GCATTG--CTATTGTTATTCCTGGCAGACCG------AGAGAGAG--AGAG--AGC-------------------------------------------------AGGCTTGGC------------------------------CT--TCTTCTT |
| droPer1 | super_1:7324752-7324918 - | LASTZ | 1 | GAG----------TGT------CGC-AAAAGTG--TTGCATACTTTTTGGCAGGCAGCAA------------------T-GCTA-AAGAGGACAG--A----GCCTTTGATTGGTTTTTCC-TTGGGCTCGGGGCTCCCAC---GT-TCTTTG------GCATTG--CTATTGTTATTCCTGGCAGACCG------AGAGAGAG--AGAGACAGC-------------------------------------------------AGGCATGGC------------------------------CT--TCTTCTT |
| droWil1 | scaffold_180708:9979069-9979243 + | LASTZ | 5 | A---------------------------AAGTG--TAGCATACTTACTGGCAGGCAAAAG---------CA-------GTGCAG-AAGAAGCTGCCACAT--TTTTTAGATTGGTTTTACG-AGT----------------TT----TGTG--------TCCTTG--C--TTGTTATTGTT---TGACTG------AAT-------------------------------------------GGTTTATTTTGTATCCCGTTTT-----GAAAGGGTTCTTTTTTTTTTGGCTCTCGTCGCCCCC-GTGTTTGC |
| droVir3 | scaffold_12963:17362502-17362582 + | LASTZ | 5 | CAG------------------------AAAGTG--TAGCATACTTTTTGGCAGTCG----TTGCCAAAGCAGCAGGCGGCGGCA-AGCAGAAGCA-------ACCTTTGATTGGTTTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 | scaffold_6500:22410643-22410826 + | LASTZ | 3 | CTGCTCCTGCCGCTGCTGCTGTCGCAGAAAGTG--TAGCATACTTTTTGGCAGTCG----TCGCAAAAGCAGCAGGC---GGCAAGGCAGCAGCA-------ACCTTTGATTGGTTTTTGG-GTT----------------T-----TGTATG------CAGTGC--TTGTTGTTATTTTC---------AGTTTGGGCAAGGGCTAAGG--------------------------------GCTCCATT-----GAAG-----A-GCTTTG-------------------------------CA--GTTTCAA |
| droGri2 | scaffold_15252:14795099-14795283 + | LASTZ | 4 | ACT----------GTTTCTCTGCTC-CTTGTCC--TTAGATACTTTTTGGCAGAGTTGCGTCGCC--------A----G-GCAA-AAGCAAAAGC--A----ACCTTTGATTGGTTTTTGG-GGT----------------T-----TCT-GT------CTGTCT--------CTCTGTCC---------GACTGT-CCGACTG--TCTC--CTCTGGTGTGGCTCTGTCTGCAATTGTGTTGTTTCATGTCGTTTAAGGGCTT-----CAAA------------------------------C---------- |
| Species | Alignment |
| dm3 | AAA----------AGTTCCTTGCGC-CAAAGTGTGTTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCTGATTGGTTTTCTT-TTTGAT-------------T---------TTCGTTCGCCATAG--GTATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TTGA------------------------------CT-GCGTTCTA |
| droSim1 | AAA----------AGTTCCTTGCGC-AAAAGTGTGTTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCTGAT----------------------------------------------------AG--ATATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TTGA------------------------------CT-GCGTTGTA |
| droSec1 | AAA----------AGTTCCTTGCGC-AAAAGTGTGTTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTGTGAT----------------------------------------------------AG--ATATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TGGA------------------------------CT-GCGGTGTA |
| droYak2 | AAA----------AGTTCTTTTCGC-AAAAGTG--TTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCAGATTGGTTTACTT-TTC----------------T-----CCTTTG------CCATAG--GTTTTGTTATTCTC---------GATTCT-------------------------------------------------------------------------TCGA------------------------------CT-GCCTTGTA |
| droEre2 | AAC----------AGTTCTTTTCGC-AAAAGTG--TTGCATACTTTTTGGCAGGCGGAAA------------------TCGCTA-AAGAGAACAA-------CCCTCAGATTGGTTTTCTT-TTG----------------C-----CCCTTG------CCATAG--GTATAGTTATTCTC---------GATTCC-------------------------------------------------------------------------TTGA------------------------------CT-GCGTTGTA |
| droEug1 | AAA----------GTTTTCTTTTGC-GAAAGTG--TTGCATACTTTTCGGCTGGCGGAAA------------------TCGCTA-AAGAGAACAA--C----CCCTCTGATTGGTTTTCTT-TCT----------------T-TGTTCTTTTG------CCATTG--CTATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TAGA------------------------------CT-GGTTTTTT |
| droBia1 | AAA----------AGTTG--TTCGC-AAAAGTG--TTGCATACTTT-TGGGAGGCGGAAA------------------TCGCTA-AAGAGAACAA-------CCCTCTGATTGGTTTTCTT-TTC----------------CCT---ACTTTT------CTACTG--CTATTGTTATTCTT---------GATTCT-------------------------------------------------------------------------TAGA------------------------------CT-GGGCTGTA |
| droTak1 | CAA----------AGTTTTTTCCAC-AAAAGTG--TTGCATACTTTTTGGGAGGCGGAAA------------------TCGCTA-AAGAGGACAG--CAA--CCCCCTGATTGGTTTCCCTTTTC----------------T-----CTTTTC------CTATTG--CTATTGTTATTTTT---------GATTCT-------------------------------------------------------------------------TCAA------------------------------CT-GGGCTGTA |
| droEle1 | AAA----------AGTTTC-ATTGC-AAAAGAG--TGGCAT---TTTAGGCAGGCGGATA------------------TCGCTA-GAGAGGAAAA-------A------ATTGTTTTTCTT-TT-------------------------TTTG------CTATAA--CTATTGTTTTTCTT---------GATTCT-------------------------------------------------------------------------TATA------------------------------T---------- |
| droRho1 | AAA----------AGTTGT-TTCGC-AAAAGTG--TTGCATATTTTTTGGCAGGCGGATA------------------TCGATA-AAGAGGACAA-------CCTTCTT---------C---TTC----------------T-----CTTTTG------CTATTG--CTAATGATATTCT--------------------------------------------------------------------------------------------A------------------------------CT-GTAATTTA |
| droFic1 | AAA----------GTTTTCCTTCGC-AAAAGTG--TTGCATACTT-TTGGCAGGCGGAAA------------------TCGCTA-AAGAGGACAA-------CCCTCTGATTGGTTTTCTT-TTT----------------C-----TTTTTG------CTGTTCCTCTATTCTTATTTTC---------GATTCG-------------------------------------------------------------------------TCGT------------------------------CT-GTGGTGTA |
| droKik1 | CGT----------TTTACCACCCAC-CAAAGTG--TTGCATACTTTTTGG-------------------------------CTA-AGAAGGACAA-------ACCTCTGATTGGTTTTCTT-TGG----------------CCT---CGT------------TCG--ACATTGTTATTCCT---------GATTCT-------------------------------------------------------------------------TCGA------------------------------CT-GGGGTGTA |
| droAna3 | ACC----------CGTTTG-CCTGC-AAAAGTG--TTGCATACTTTTTGGCAGGCTAAC-------------------G--------GGGGACAG--ACCTCCCCTGCGATTGGTTTTCTT-CTC----------------T---------TT------CATCCG--CCATTGTTATTCCT---------GATTCG-------------------------------------------------------------------------GAGA------------------------------GTCGCATTCGA |
| droBip1 | ACC----------CGTTTT-CCGGC-AAAAGTG--TTGCATACTTTTTGGCAGGC---------------------------TA-ACG-GGACAG-------ACCTCCGATTGGTTTTCTT-TTC----------------T---------TT------CATCCG--CCATTGTTATTCCT---------GATTCG-------------------------------------------------------------------------GGGA------------------------------AT-------CA |
| dp4 | GAG----------TGT------CGC-AAAAGTG--TTGCATACTTTTTGGCAGGCAGCAA------------------T-GCTA-AAGAGGGCAG--A----GCCTTTGATTGGTTTTTCT-TTGGGCTCGGGGCTCCCAC---GT-TCTGTG------GCATTG--CTATTGTTATTCCTGGCAGACCG------AGAGAGAG--AGAG--AGC-------------------------------------------------AGGCTTGGC------------------------------CT--TCTTCTT |
| droPer1 | GAG----------TGT------CGC-AAAAGTG--TTGCATACTTTTTGGCAGGCAGCAA------------------T-GCTA-AAGAGGACAG--A----GCCTTTGATTGGTTTTTCC-TTGGGCTCGGGGCTCCCAC---GT-TCTTTG------GCATTG--CTATTGTTATTCCTGGCAGACCG------AGAGAGAG--AGAGACAGC-------------------------------------------------AGGCATGGC------------------------------CT--TCTTCTT |
| droWil1 | A---------------------------AAGTG--TAGCATACTTACTGGCAGGCAAAAG---------CA-------GTGCAG-AAGAAGCTGCCACAT--TTTTTAGATTGGTTTTACG-AGT----------------TT----TGTG--------TCCTTG--C--TTGTTATTGTT---TGACTG------AAT-------------------------------------------GGTTTATTTTGTATCCCGTTTT-----GAAAGGGTTCTTTTTTTTTTGGCTCTCGTCGCCCCC-GTGTTTGC |
| droVir3 | CAG------------------------AAAGTG--TAGCATACTTTTTGGCAGTCG----TTGCCAAAGCAGCAGGCGGCGGCA-AGCAGAAGCA-------ACCTTTGATTGGTTTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 | CTGCTCCTGCCGCTGCTGCTGTCGCAGAAAGTG--TAGCATACTTTTTGGCAGTCG----TCGCAAAAGCAGCAGGC---GGCAAGGCAGCAGCA-------ACCTTTGATTGGTTTTTGG-GTT----------------T-----TGTATG------CAGTGC--TTGTTGTTATTTTC---------AGTTTGGGCAAGGGCTAAGG--------------------------------GCTCCATT-----GAAG-----A-GCTTTG-------------------------------CA--GTTTCAA |
| droGri2 | ACT----------GTTTCTCTGCTC-CTTGTCC--TTAGATACTTTTTGGCAGAGTTGCGTCGCC--------A----G-GCAA-AAGCAAAAGC--A----ACCTTTGATTGGTTTTTGG-GGT----------------T-----TCT-GT------CTGTCT--------CTCTGTCC---------GACTGT-CCGACTG--TCTC--CTCTGGTGTGGCTCTGTCTGCAATTGTGTTGTTTCATGTCGTTTAAGGGCTT-----CAAA------------------------------C---------- |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
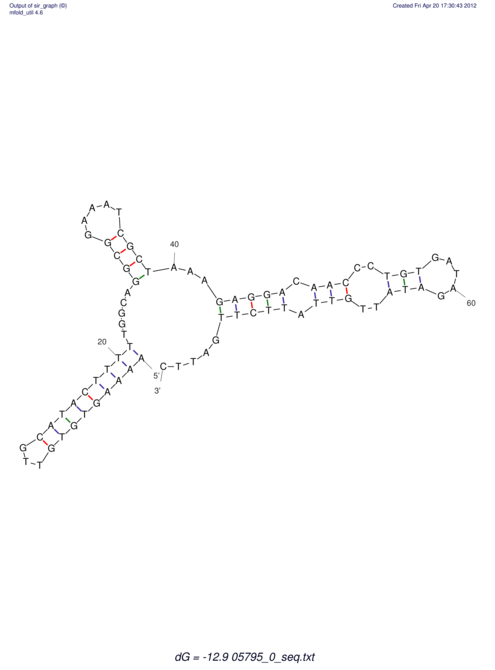
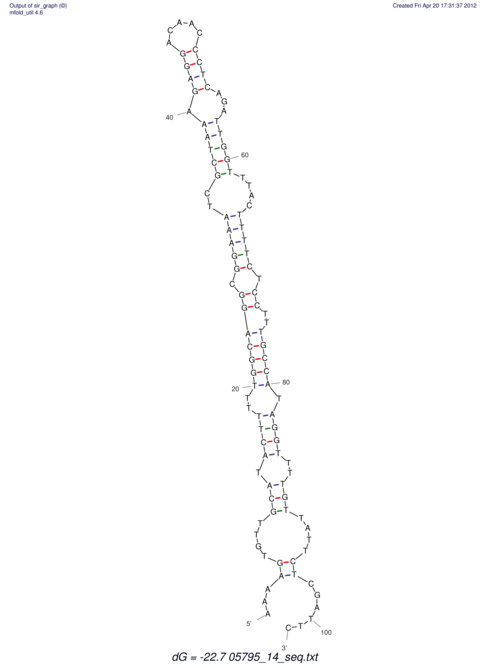
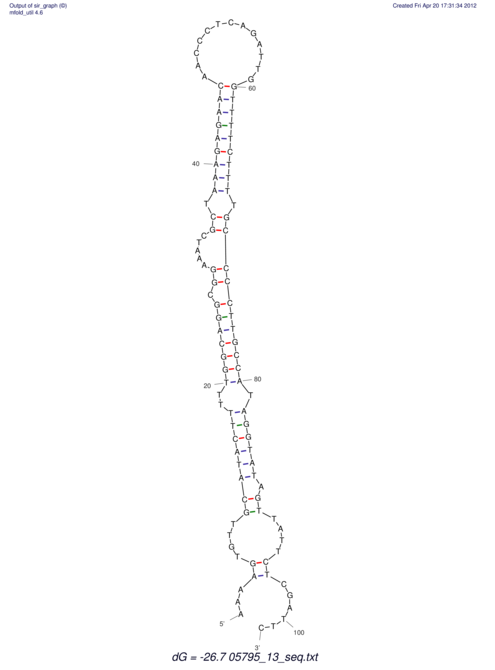
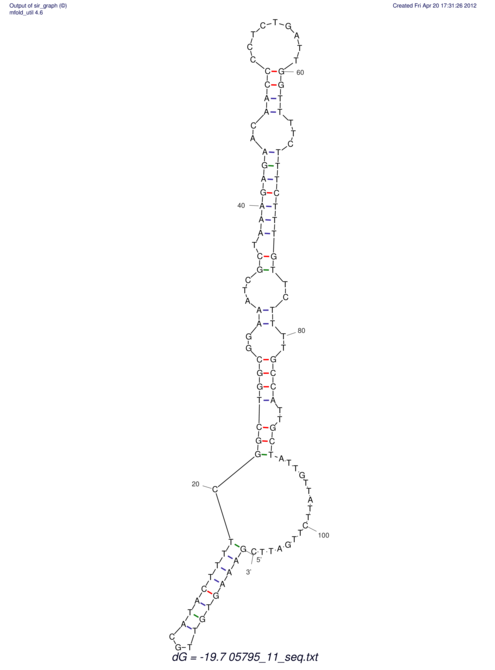
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:31:58 EDT