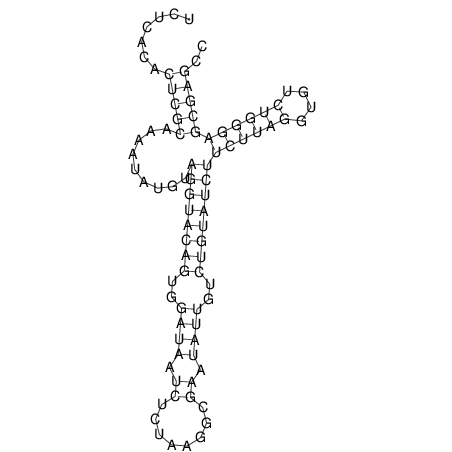
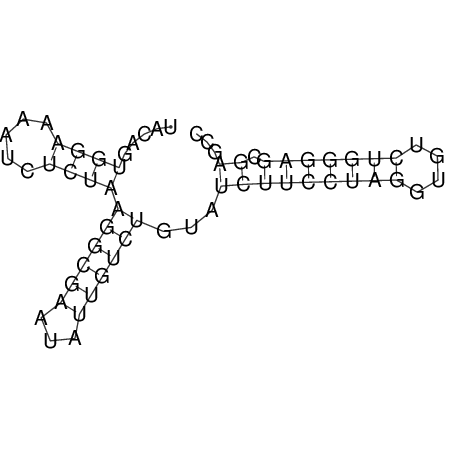
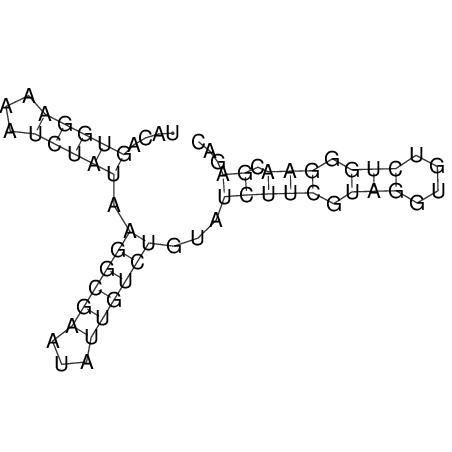
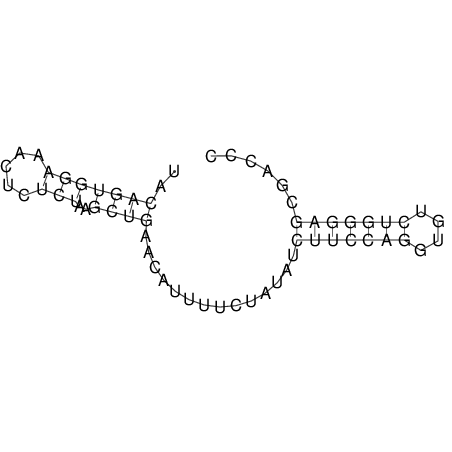
| dm3 |
chr2L:2765167-2765282 - |
NA |
TTTGTGTAACCATTATCTCACAC--------------TCGCAAAATATGTAGGTA-------CAG-----------------------------------------------------------------------------------------TGGATAATCTCTAAGGCGAATAT-TGTCTGT-ATC---------------------TTCTTAGGTGTCTGGGAGCGAGCCACA---AGTGCTGA---GTAT |
| droSim1 |
chr2L:2727612-2727727 - |
2 |
TTTGTGTAACCATTACCTCACAC--------------TCGCCAAATATGAAGGTA-------CAG-----------------------------------------------------------------------------------------TGGAAAATCTCTAAGGCGAATAT-TGTCTGT-ATC---------------------TTCCTAGGTGTCTGGGAGCGAGCCACA---AGTGCGGA---GTAT |
| droSec1 |
super_5:931138-931253 - |
1 |
TTTGTGTAACCATTTTCTCACAC--------------TCGCCAAATATGAAGGTA-------CAG-----------------------------------------------------------------------------------------TGGAAAATCTATAAGGCGAATAT-TGTCTGT-ATC---------------------TTCGTAGGTGTCTGGGAACGAGACACA---AGTGCGGA---ATAT |
| droYak2 |
chr2L:2760749-2760863 - |
1 |
TTTGTATAATCATTATCTTACAT--------------TCAAATAATATGTAGGTA-------CAG-----------------------------------------------------------------------------------------TGGAAACTCTCTAAGCTGAACAT-TTTCTA-TA-T---------------------CTTCCAGGTGTCTGGGAGCGACCCACT---ACTGCGGA---GTAT |
| droEre2 |
scaffold_4929:2817993-2818131 - |
1 |
TAAAACC----ATAATCTCACAT--------------TTGCATAATATGTAGGTA-------CA-----------------------------------------TGGA------------AAAACGATCATCA---------GTAGGTCACAGTGGAAACTCTCTAAGACGAATGT-GGTCTAT-ATC---------------------TTCCTAGGTGTCTGGGAGCGACCCGCT---AGTGCTGA---ATAC |
| droEug1 |
scf7180000409554:3346024-3346246 + |
1 |
TTTGTATAATCATAATTTCCCATGACTATATTTCAATGCCCCTATTGTGGGGCTT-------CAG----TACA-CATCTTCA---AGAATCATTTAATATTCACATTGGGATGCAGATTATCATATAATCATCACCTTAGTGTGTAAAGGACAATGAAAGCTT-CCAAAAAAAATTT---TTTA-TT-T--------AAGTTTTTCCATCTTTTTAGGTGTCTGGGAGCGGCCAACAATGAGTTCCGA---GTAC |
| droBia1 |
scf7180000302188:1767970-1768092 - |
1 |
TTTGTATAAG-GTCTA-----------------------------------AGGACATTGGCCGTTAT-TAA--C--CATAT---AAA--------------------------------------------------------------ACTATAAAAATTACATAACTTAAATAT-ACTTATT-TCC---------------------TGTTTAGGTTTCTGGGAGCGACCCACAACCAGTGCCGA---GTAC |
| droTak1 |
scf7180000415705:514059-514162 - |
1 |
ATTCATTGAAT------------------------------------------CCATTCATGCAGTGCTCTA--A--TATGTATAG--------------------------------------------------------------------CAAATAAT-----------ATATATTTTTG-TA-C---------------------CTTTTAGGTTTCTGGGAGCGACCAACGTCTAGCGCCGA---GTAC |
| droEle1 |
scf7180000491273:412313-412354 + |
2 |
TTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGGTGTCTGGGAGCGACCCACGTCTAATGCCGA---CTTC |
| droRho1 |
scf7180000766156:59158-59229 + |
2 |
CTAAAGCAACATTTATTA----T---------------------------------------------------------------------------------------------------------------------------------------------------------T-TGTTTCAAA-A---------------------CTTGTAGATGTCTGGGCGCGACCCACAACAAATGCCGA---GTAC |
| droFic1 |
scf7180000453924:716599-716727 - |
1 |
TATGTACATATGGA-------------------------------------T-----GAGTATA----------C--TTTTT---AAAATTAAAAAATATATAAATTGG------------TTTA-----------------------------TGAATATCATCTCA-ATAAATAC-TTTTTG-AA-C---------------------TTTTTAGGTGTCTGGGAGCGACCCACAACGAATTCCGA---GTAT |
| droKik1 |
scf7180000302271:228107-228177 - |
1 |
TTTCTAAAACC---------------------------------------------------------------------------------------------------------------------------------------------------------------TCAACAA-ATTATG-T--A---------------------TTTATAGGAGTCTGGGAGCGACCCTCATCGAGTACCGAAACCTTT |
| droAna3 |
scaffold_12984:244381-244458 - |
2 |
TAC------------------------------------------------------------AGTGGGAACG--------------------------------------------------------------------------------------AGTCTTAAAAATGAAAAA-T---------A---------------------TTTTCAGGCATCTGGGAGCGACCAGCACAGACGGCCACTTCTTAT |
| droBip1 |
scf7180000396574:369645-369692 - |
2 |
AT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A---------------------CTTTCAGGTATCTGGGAGCGACCAGCACAGACGGCCGCTTCTTAT |
| dp4 |
chr4_group4:2108184-2108280 - |
1 |
TTTGACGCTA-ATTGG-----------------------------------TGCACACTAAGCAGTGGGTCCGG-------TCTGA------------------------------------------------------------------------------------CAAA-----TGAAG-TT-T---------------------TCCGCAGGTTTCTGGGAGCGCCCCACATCGTCCACCAC---GTAT |
| droPer1 |
super_10:1114845-1114941 - |
1 |
TTTGACGCTA-ATTGG-----------------------------------TGCACACTAAGCAGTGGGTCCGG-------TCTGA------------------------------------------------------------------------------------CAAA-----TGAAG-TT-T---------------------TCCGCAGGTTTCTGGGAGCGCCCCACATCGTCCACCAC---GTAT |
| droWil1 |
scaffold_180772:1542185-1542212 - |
1 |
CT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTGGGAACGACCAGC------CACAAA---ATAT |
| droVir3 |
scaffold_12963:11387645-11387742 - |
1 |
TGCGTATGCTAGATA------------------------------------------CC-AGAAGCAGATCAAG-------TTTG-------------------------------------------------------------------------------CTCA-CTTTACAT-TTTATG-TC-C---------------------GCAACAGCTTTCTGGGAGAGTTTTAATCCAGTTAAAAG---ATTT |
| droMoj3 |
scaffold_6500:20137581-20137687 - |
1 |
TGAGCTA----ATTGG-----------------------------------AATCCAAAGTGCAGTTTTTTA--T--ATTATATAT--------------------------------------------------------------------ACAT---------A-CAT-ATATATATATG-TA-T---------------------ATTTAAGCGTTCTGGGAGAGCTACAATTCAGCTAAAAA---GTTT |
| droGri2 |
scaffold_15126:5047932-5047984 - |
1 |
AATATTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAA-TT-CCCAATAGC-------------TTGCTGGA---AGAAGAGCTTTCCACA--------TAG---GTTT |