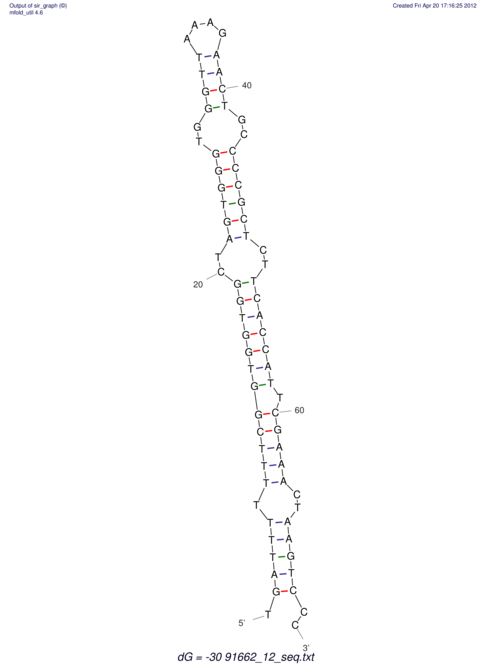
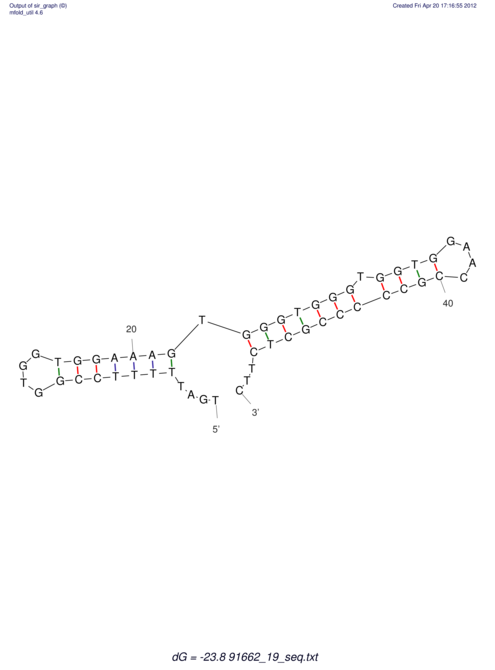
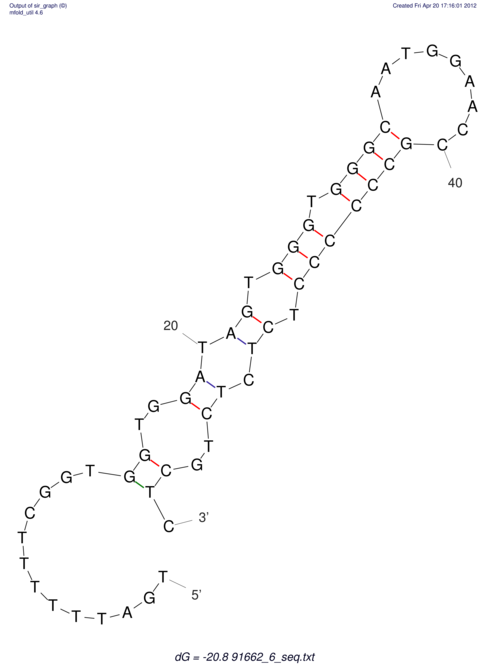
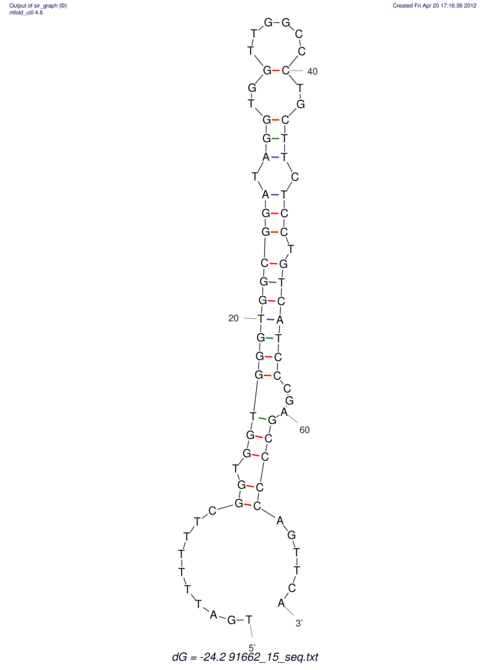
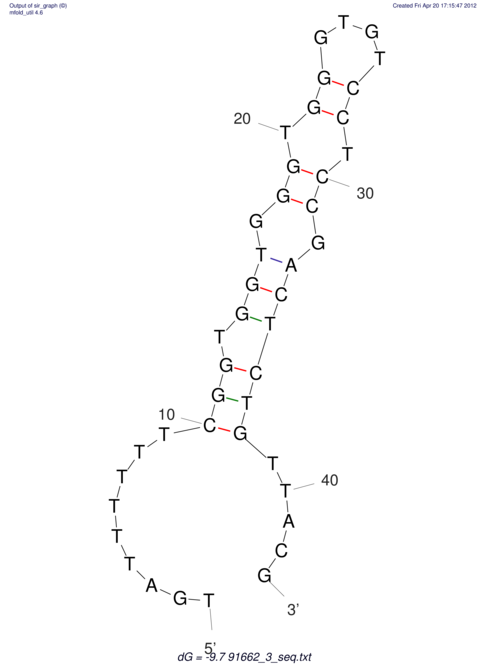
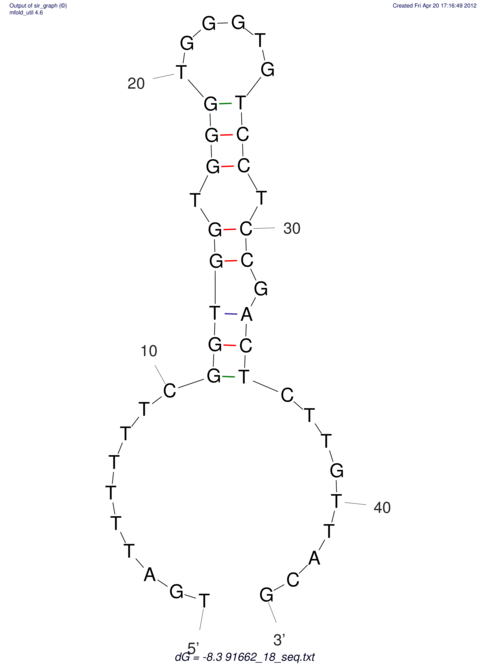
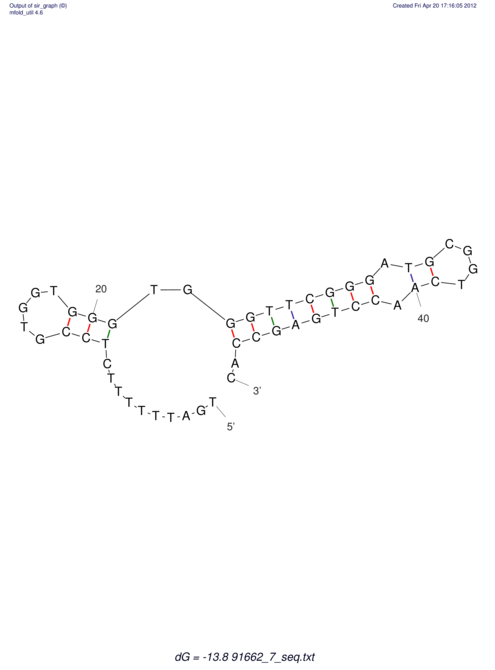
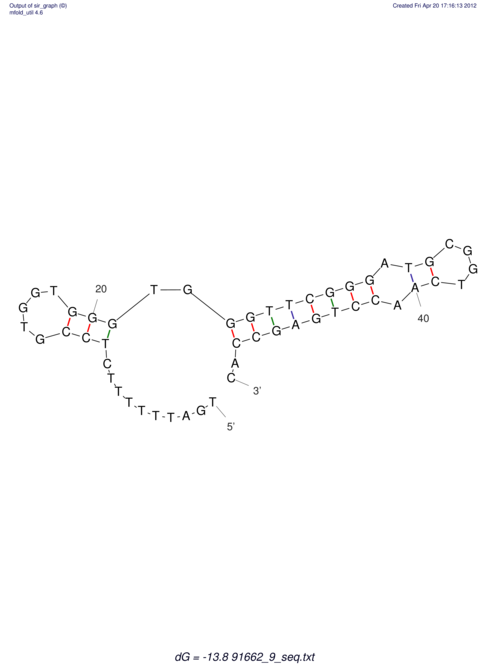
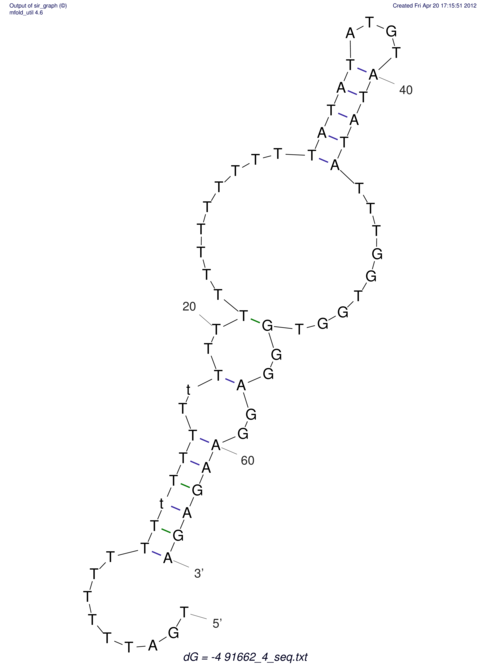
| TTTGCTGTT-TTGCGATGCTCTATTGATTTTTTTTtTTTTtTTTTTTTTTTTTTTATATATGTATATATTTGGTGGTGG---------------------GAGGAAGAGA----------------------------------------------------------------------------------------AG | Size | Hit Count | Total Norm | V117
Male body | V118
Embryo | V119
Head | M020
Head |
| .............................TTTTTTTTTTTTTTTTTTT........................................................................................................................................................ | 19 | 89 | 0.08 | 0.00 | 0.08 | 0.00 | 0.00 |
| .................................ATTTTTTTTTTTTTTTTTTTTT................................................................................................................................................. | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| .............................ATTTTTTTTTTTTTTTTTTTTT..................................................................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ...........................ATTTTTTTTTTTTTTTTTTTTT....................................................................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ..............................ATTTTTTTTTTTTTTTTTTTTT.................................................................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ................................ATTTTTTTTTTTTTTTTTTTTT.................................................................................................................................................. | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ...............................ATTTTTTTTTTTTTTTTTTTTT................................................................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ............................ATTTTTTTTTTTTTTTTTTTTT...................................................................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ..........................ATTTTTTTTTTTTTTTTTTTTT........................................................................................................................................................ | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ..................................TTTTTTTTTTTTTTTTTTT................................................................................................................................................... | 19 | 89 | 0.04 | 0.00 | 0.00 | 0.04 | 0.00 |
| ............................TTTTTTTTTTTTTTTTTTTTTT...................................................................................................................................................... | 22 | 58 | 0.07 | 0.00 | 0.00 | 0.07 | 0.00 |
| ...........................ATTTTTTTTTTTTTTTTTTTT........................................................................................................................................................ | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ................................TTTTTTTTTTTTTTTTTTTTTTT................................................................................................................................................. | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ..........................TTTTTTTTTTTTTTTTTTTTTTT....................................................................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ..............................TTTTTTTTTTTTTTTTTTTTTTT................................................................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| .............................ATTTTTTTTTTTTTTTTTTTTTTTT.................................................................................................................................................. | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ..............................TTTTTTTTTTTTTTTTTTTTT..................................................................................................................................................... | 21 | 68 | 0.04 | 0.00 | 0.00 | 0.00 | 0.04 |
| ...............................TTTTTTTTTTTTTTTTTTTTTTT.................................................................................................................................................. | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ..........................ATTTTTTTTTTTTTTTTTTTTTTTT..................................................................................................................................................... | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ..................................ATTTTTTTTTTTTTTTTTTTT................................................................................................................................................. | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .................................TTTTTTTTTTTTTTTTTTTTTTT................................................................................................................................................ | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ............................ATTTTTTTTTTTTTTTTTTTT....................................................................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ................................ATTTTTTTTTTTTTTTTTTTT................................................................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ..............................ATTTTTTTTTTTTTTTTTTTTTTTT................................................................................................................................................. | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ............................ATTTTTTTTTTTTTTTTTTTTTTTT................................................................................................................................................... | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ...............................ATTTTTTTTTTTTTTTTTTTT.................................................................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ..............................ATTTTTTTTTTTTTTTTTTTT..................................................................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ..........................ATTTTTTTTTTTTTTTTTTTT......................................................................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .................................ATTTTTTTTTTTTTTTTTTTT.................................................................................................................................................. | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ...........................ATTTTTTTTTTTTTTTTTTTTTTTT.................................................................................................................................................... | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ............................TTTTTTTTTTTTTTTTTTTTTTT..................................................................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| .............................ATTTTTTTTTTTTTTTTTTTT...................................................................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .............................TTTTTTTTTTTTTTTTTTTTTTT.................................................................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ...........................TTTTTTTTTTTTTTTTTTTTTTT...................................................................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ..................................TTTTTTTTTTTTTTTTTTTTTTT............................................................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |