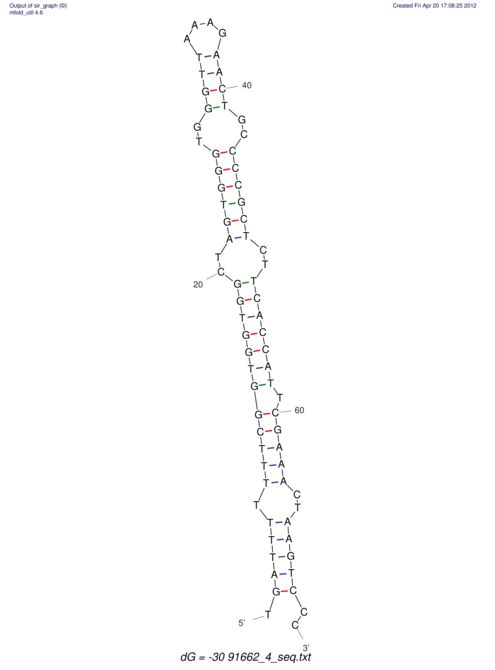
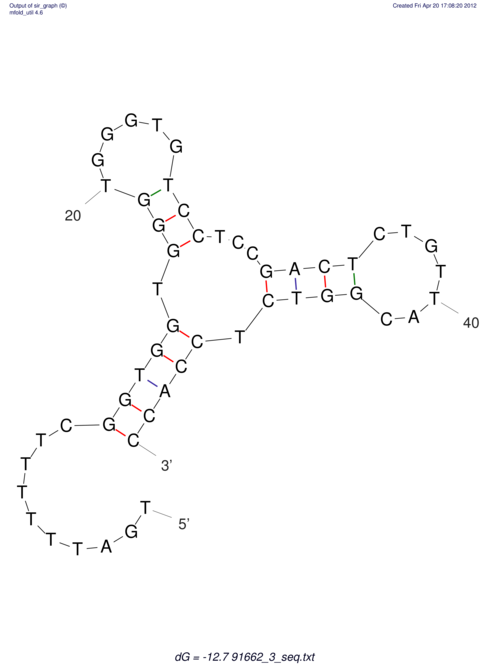
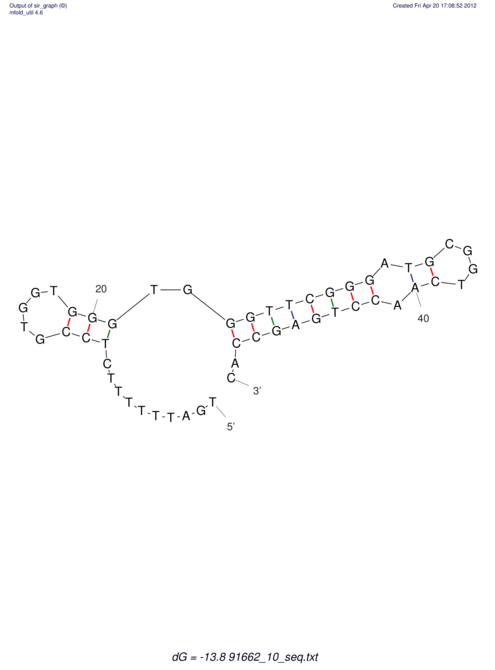
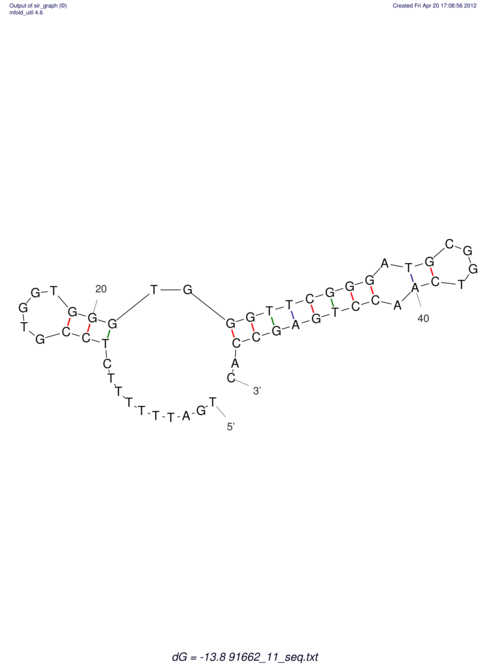
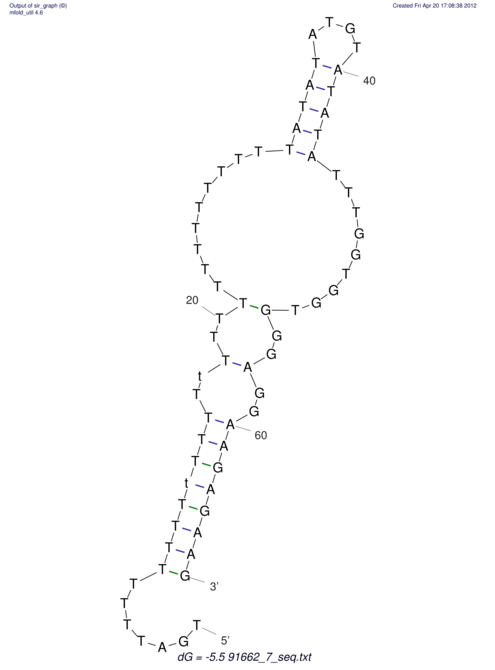
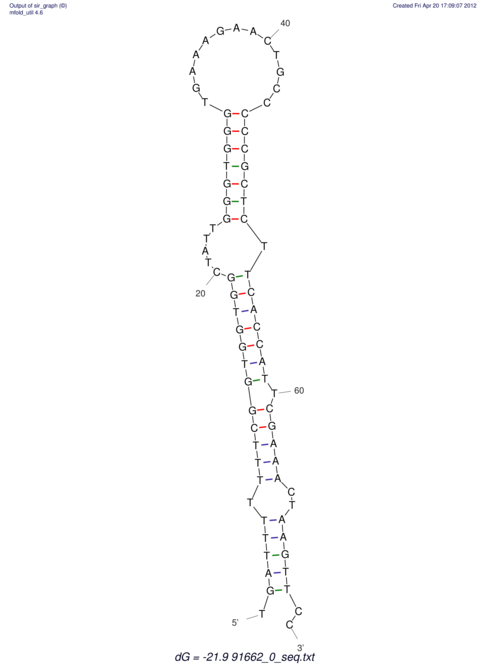
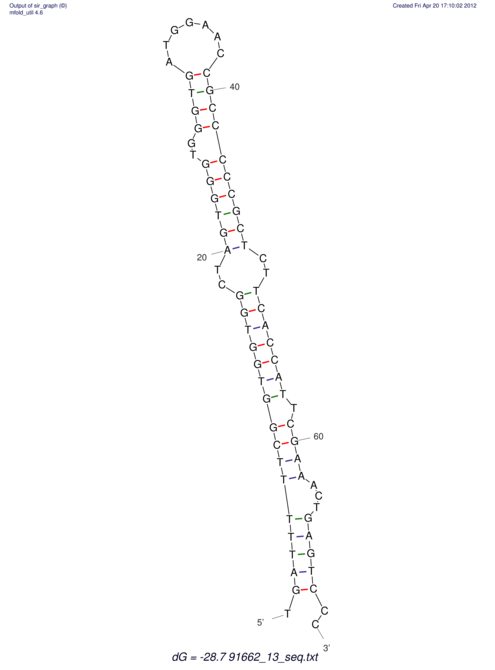
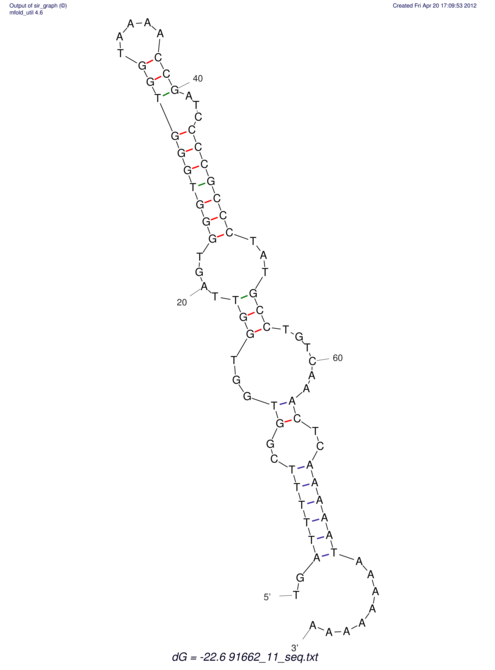
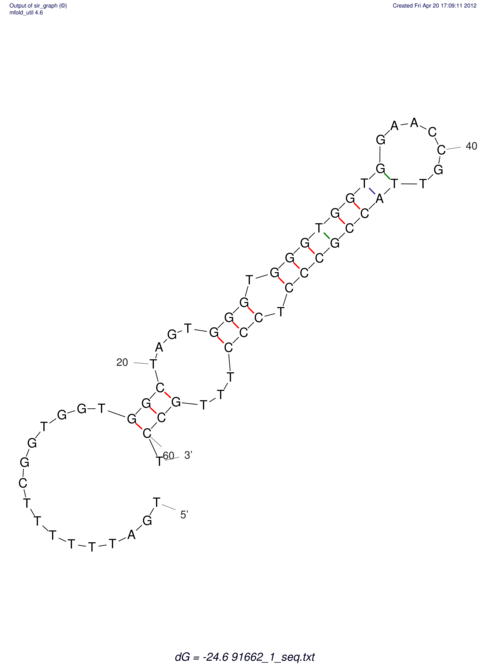
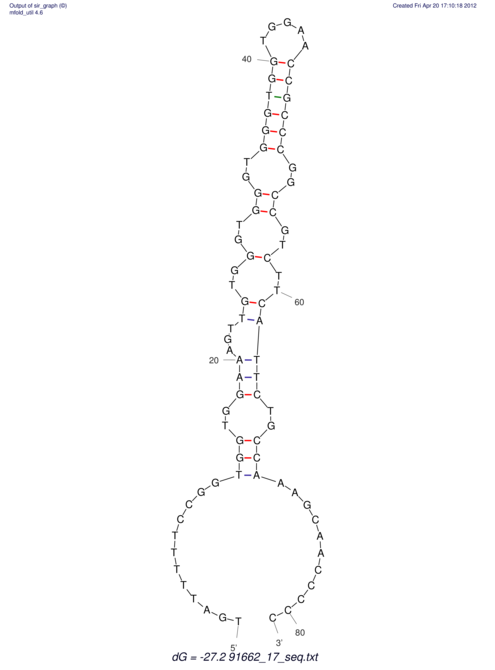
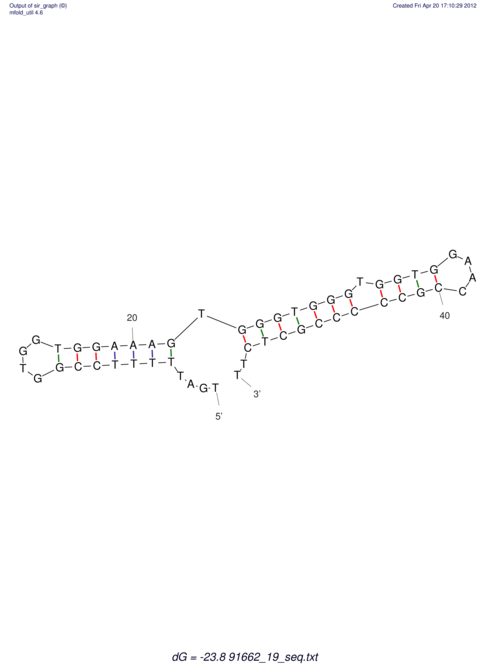
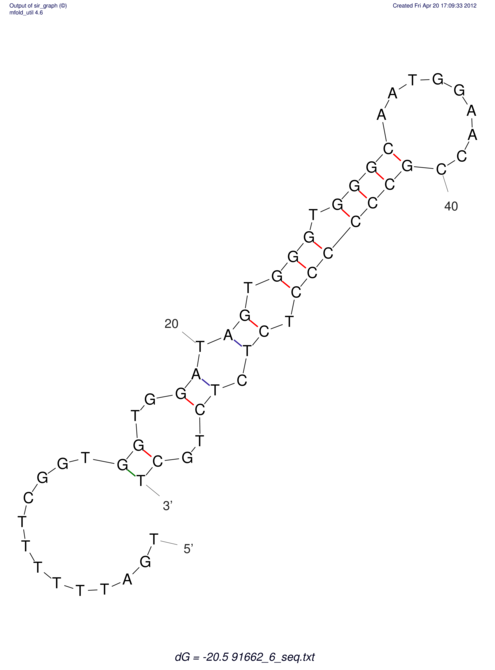
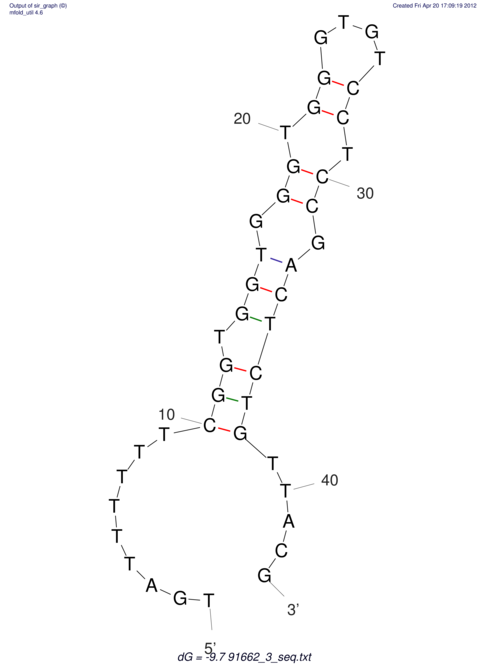
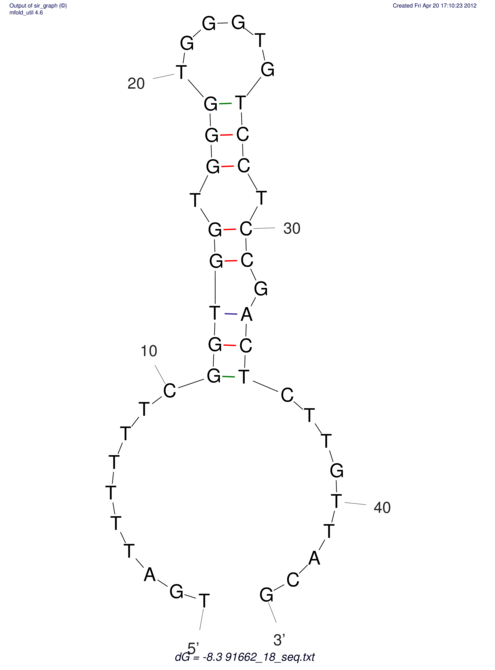
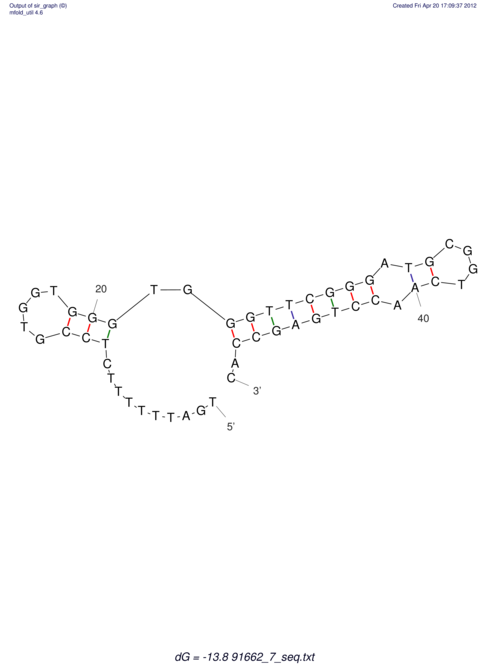
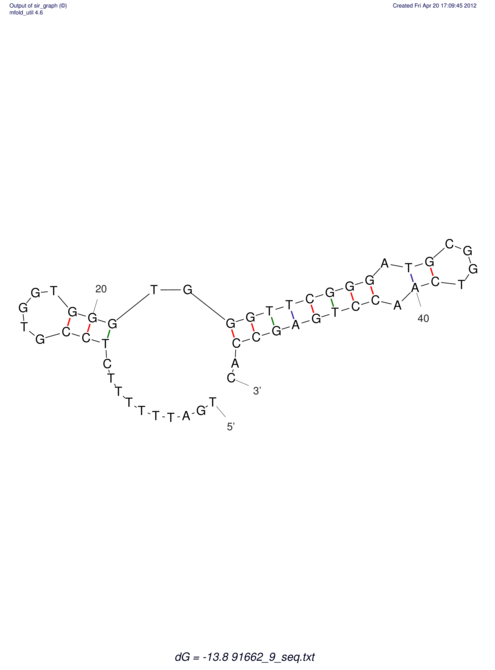
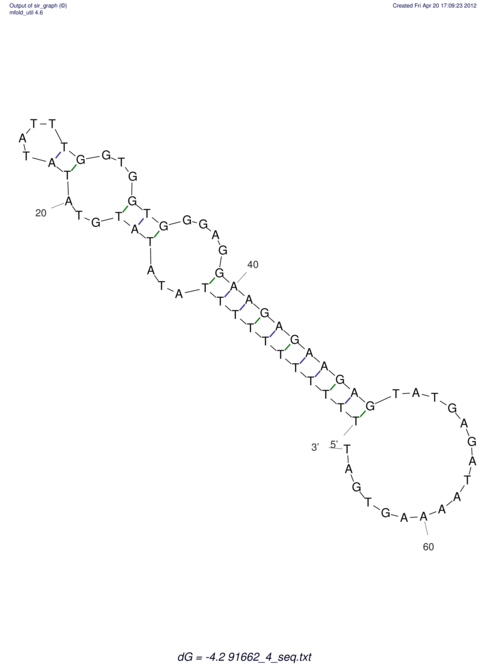
| TTTGCTGTTTTGCGATGCTCTATTGATTTTTT-TTtTTTTtTTTTTTTTTTTTTTATATATGTATATATTTGGTGGTG-------GGAGGAA------GAGA-----AG------------------------------------------------- | Size | Hit Count | Total Norm | V117
Male body | V118
Embryo | V119
Head | M020
Head |
| ............................TTTT-TTTTTTTTTTTTTTT.............................................................................................................. | 19 | 89 | 0.08 | 0.00 | 0.08 | 0.00 | 0.00 |
| .................................ATTTTTTTTTTTTTTTTTTTTT....................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ............................ATTT-TTTTTTTTTTTTTTTTTT........................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ..........................ATTTTT-TTTTTTTTTTTTTTTT............................................................................................................. | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| .............................ATT-TTTTTTTTTTTTTTTTTTT.......................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ...............................A-TTTTTTTTTTTTTTTTTTTTT........................................................................................................ | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ..............................AT-TTTTTTTTTTTTTTTTTTTT......................................................................................................... | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ...........................ATTTT-TTTTTTTTTTTTTTTTT............................................................................................................ | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| .........................ATTTTTT-TTTTTTTTTTTTTTT.............................................................................................................. | 22 | 16 | 0.31 | 0.00 | 0.31 | 0.00 | 0.00 |
| ..................................TTTTTTTTTTTTTTTTTTT......................................................................................................... | 19 | 89 | 0.04 | 0.00 | 0.00 | 0.04 | 0.00 |
| ...........................TTTTT-TTTTTTTTTTTTTTTTT............................................................................................................ | 22 | 58 | 0.07 | 0.00 | 0.00 | 0.07 | 0.00 |
| ..........................ATTTTT-TTTTTTTTTTTTTTT.............................................................................................................. | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ...............................T-TTTTTTTTTTTTTTTTTTTTTT....................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| .........................TTTTTTT-TTTTTTTTTTTTTTTT............................................................................................................. | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| .............................TTT-TTTTTTTTTTTTTTTTTTTT......................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ............................ATTT-TTTTTTTTTTTTTTTTTTTTT........................................................................................................ | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| .............................TTT-TTTTTTTTTTTTTTTTTT........................................................................................................... | 21 | 68 | 0.04 | 0.00 | 0.00 | 0.00 | 0.04 |
| ..............................TT-TTTTTTTTTTTTTTTTTTTTT........................................................................................................ | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| .........................ATTTTTT-TTTTTTTTTTTTTTTTTT........................................................................................................... | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ..................................ATTTTTTTTTTTTTTTTTTTT....................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .................................TTTTTTTTTTTTTTTTTTTTTTT...................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ...........................ATTTT-TTTTTTTTTTTTTTTT............................................................................................................. | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ...............................A-TTTTTTTTTTTTTTTTTTTT......................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .............................ATT-TTTTTTTTTTTTTTTTTTTTTT....................................................................................................... | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ...........................ATTTT-TTTTTTTTTTTTTTTTTTTT......................................................................................................... | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ..............................AT-TTTTTTTTTTTTTTTTTTT.......................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .............................ATT-TTTTTTTTTTTTTTTTTT........................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .........................ATTTTTT-TTTTTTTTTTTTTT............................................................................................................... | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| .................................ATTTTTTTTTTTTTTTTTTTT........................................................................................................ | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ..........................ATTTTT-TTTTTTTTTTTTTTTTTTT.......................................................................................................... | 25 | 10 | 0.30 | 0.00 | 0.30 | 0.00 | 0.00 |
| ...........................TTTTT-TTTTTTTTTTTTTTTTTT........................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ............................ATTT-TTTTTTTTTTTTTTTTT............................................................................................................ | 21 | 19 | 0.16 | 0.00 | 0.16 | 0.00 | 0.00 |
| ............................TTTT-TTTTTTTTTTTTTTTTTTT.......................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ..........................TTTTTT-TTTTTTTTTTTTTTTTT............................................................................................................ | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ..................................TTTTTTTTTTTTTTTTTTTTTTT..................................................................................................... | 23 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |