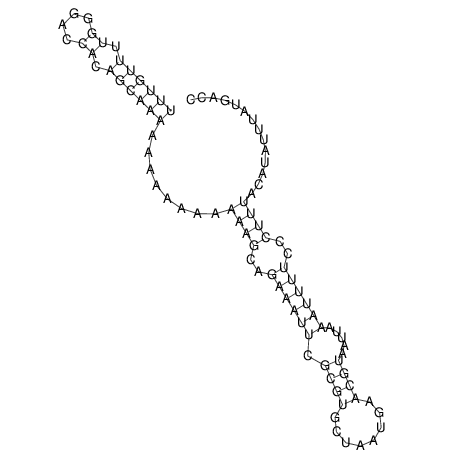
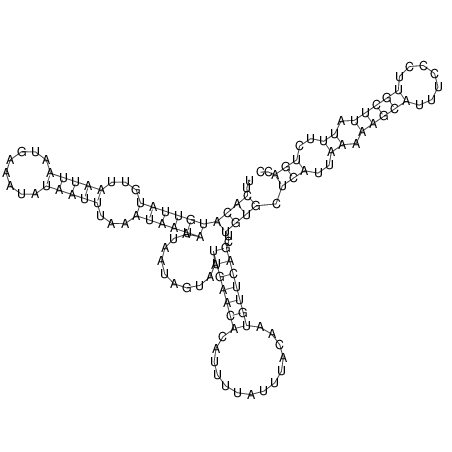
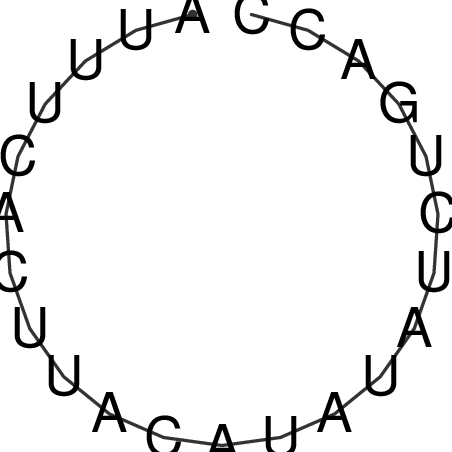
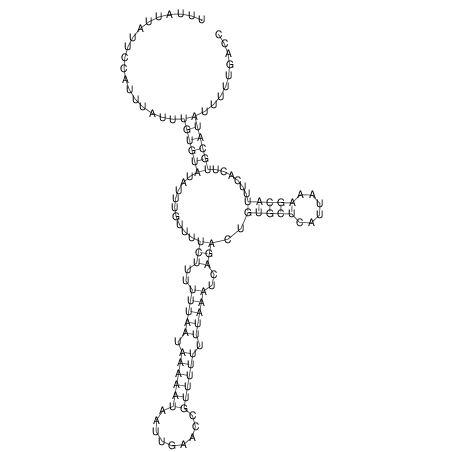
| CAGATAAGGACATT-GTGT--------------------------------TTTGTT-TTGGGACCACAGCAAAAAAAAAAAAAAG-------------------------CA-----GAAATTCGCGT--GCTA-------------------------------------ATGAACGTAATTAAATTTTC--CC-TTTACATATTT-ATGACCCAATTT----TG----TTT | Size | Hit Count | Total Norm | V117
Male body | V118
Embryo | V119
Head | M020
Head |
| ..........................................................................AAAAAAAAAGAA-------------------------CA-----GAAA................................................................................................................ | 18 | 51 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| .......................................................................AAAAAAAAAAAAAAG-------------------------AA-----GA.................................................................................................................. | 19 | 15 | 0.20 | 0.00 | 0.20 | 0.00 | 0.00 |
| ..GATAAxGACATT-GTTT--------------------------------TTT.................................................................................................................................................................................... | 18 | 1 | 2.00 | 0.00 | 2.00 | 0.00 | 0.00 |
| ........................................................................AAAAAAAAAAAAAG-------------------------AA-----GA.................................................................................................................. | 18 | 20 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 |
| .........................................................................AAAAAAAAAAGAA-------------------------CA-----GAA................................................................................................................. | 18 | 49 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| .......................................................................AAAAAAAAAAAAAGG-------------------------AA-----GA.................................................................................................................. | 19 | 40 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ........................................................................AATAAAAAAAAAAG-------------------------AA-----GAAA................................................................................................................ | 20 | 26 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| ...........................................................................AAAAAAAACAG-------------------------AA-----GAAAT............................................................................................................... | 18 | 13 | 0.08 | 0.08 | 0.00 | 0.00 | 0.00 |
| ........................................................................AAAAAAAAAAAAAG-------------------------GA-----GC.................................................................................................................. | 18 | 16 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| ....................................................................AGCAAATAAAAGAAAAAG-------------------------........................................................................................................................... | 18 | 3 | 0.33 | 0.00 | 0.33 | 0.00 | 0.00 |
| .......................................................................AAAACAAAAAAAAAG-------------------------CA-----G................................................................................................................... | 18 | 5 | 0.20 | 0.00 | 0.20 | 0.00 | 0.00 |
| ........................................................................AAAAAGAAAAAAAG-------------------------CA-----AAA................................................................................................................. | 19 | 25 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| ............................................................................................................................................................................................................ATTT-ATGAACCAATTG----TG----TT. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ........................................................................AAAAACAAAAAAAG-------------------------CA-----GGA................................................................................................................. | 19 | 3 | 0.33 | 0.00 | 0.33 | 0.00 | 0.00 |
| .................................................................AACAGCCAAAAAAAAAAA....................................................................................................................................................... | 18 | 16 | 0.06 | 0.00 | 0.06 | 0.00 | 0.00 |
| .................................................................AACAGCGAAAAAAAAAAA....................................................................................................................................................... | 18 | 18 | 0.06 | 0.00 | 0.00 | 0.06 | 0.00 |
| .........................................................................AAAAAAAAAAGAG-------------------------AA-----GAAA................................................................................................................ | 19 | 31 | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 |
| ..................................................................ATTGCAAAAAAAAAAAAA...................................................................................................................................................... | 18 | 50 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ..........................................................................AAAAAAAAAAAG-------------------------GA-----GAAAAT.............................................................................................................. | 20 | 6 | 0.17 | 0.00 | 0.17 | 0.00 | 0.00 |
| ..........................................................................AAAGAAAAAGAG-------------------------CA-----GAAATT.............................................................................................................. | 20 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| .................................................................AACAGCAAAAAAAGAAAA....................................................................................................................................................... | 18 | 10 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 |
| ...................................................................CAGCGAAAAAAAAAGAAA..................................................................................................................................................... | 18 | 6 | 0.17 | 0.00 | 0.17 | 0.00 | 0.00 |
| .............................................................................AAAAAAAAG-------------------------CA-----GAAAAAC............................................................................................................. | 18 | 14 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| .......................................................................AAAAAAAAAAACAAG-------------------------AA-----GA.................................................................................................................. | 19 | 34 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| ........................................................................AAAAACAAAAAAAG-------------------------AA-----GAA................................................................................................................. | 19 | 27 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| .......................................................................AAAAAAAAAACAAAG-------------------------AA-----GAA................................................................................................................. | 20 | 14 | 0.07 | 0.00 | 0.07 | 0.00 | 0.00 |
| ...................................................................CCCCAAAAAAAAAAAAAA..................................................................................................................................................... | 18 | 50 | 0.02 | 0.00 | 0.00 | 0.00 | 0.02 |
| .......................................................................AAAAAAAAACAAAAG-------------------------AA-----GA.................................................................................................................. | 19 | 32 | 0.03 | 0.00 | 0.03 | 0.00 | 0.00 |
| .........................................................................AAAAAAAAAAGAG-------------------------CA-----CAA................................................................................................................. | 18 | 7 | 0.14 | 0.00 | 0.14 | 0.00 | 0.00 |
| ........................................................................AAAAAAAAAAAACC-------------------------CA-----GAA................................................................................................................. | 19 | 5 | 0.20 | 0.00 | 0.20 | 0.00 | 0.00 |
| ...................................................................CAGAAAAAAAAAACAAAAG-------------------------........................................................................................................................... | 19 | 10 | 0.10 | 0.00 | 0.00 | 0.10 | 0.00 |
| .................................................................CACAGCACAAAAAAACAAAA..................................................................................................................................................... | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| ..........................................................................AAAAAAAAAAGG-------------------------AA-----GAAA................................................................................................................ | 18 | 46 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| ........................................................................AAAAACAAAAAAGG-------------------------CA-----GA.................................................................................................................. | 18 | 3 | 0.33 | 0.00 | 0.33 | 0.00 | 0.00 |
| ........................................................................AAAAAAAAAAAAAG-------------------------AA-----GACA................................................................................................................ | 20 | 25 | 0.04 | 0.00 | 0.04 | 0.00 | 0.00 |
| .......................................................................AAAAAAAAAGAAAAG-------------------------CA-----GA.................................................................................................................. | 19 | 10 | 0.10 | 0.00 | 0.10 | 0.00 | 0.00 |
| .................................................................TACAGCAAAAAAAAAAAA....................................................................................................................................................... | 18 | 22 | 0.05 | 0.00 | 0.05 | 0.00 | 0.00 |
| ................................................................CCACAGGAAGAAAAAAAA........................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................ACAGCCAAACAAAAAAAAA..................................................................................................................................................... | 19 | 3 | 0.33 | 0.00 | 0.33 | 0.00 | 0.00 |