
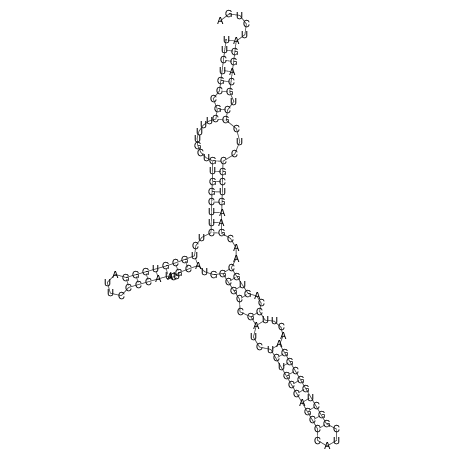
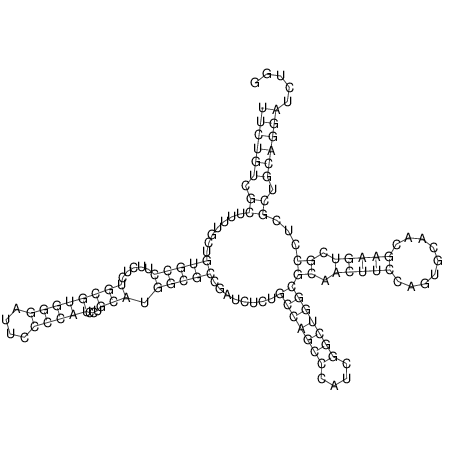
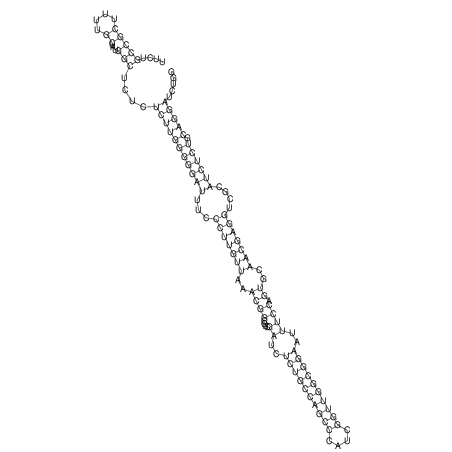
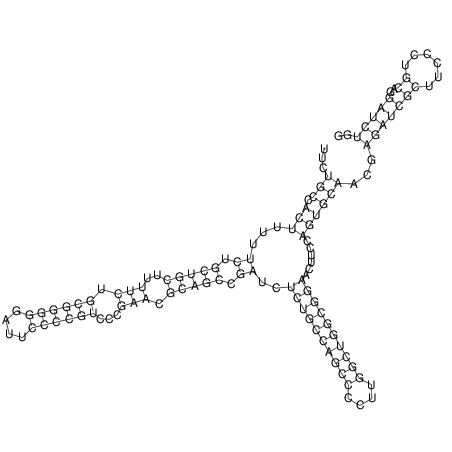
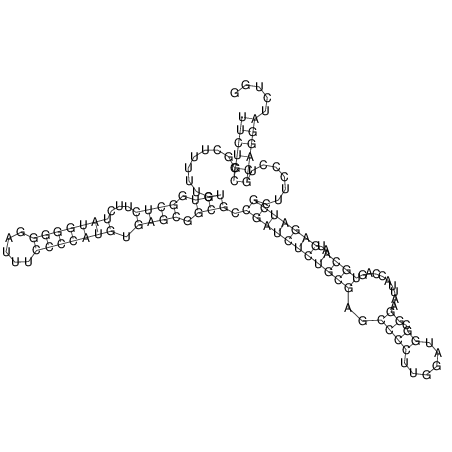
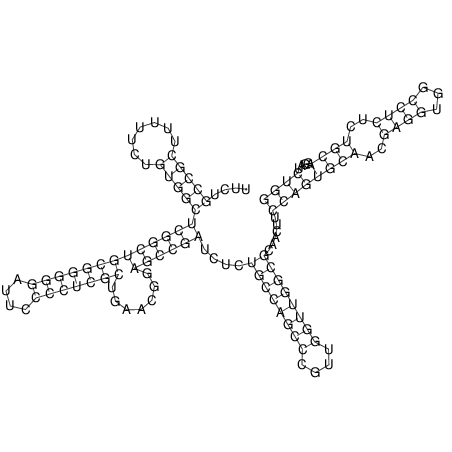
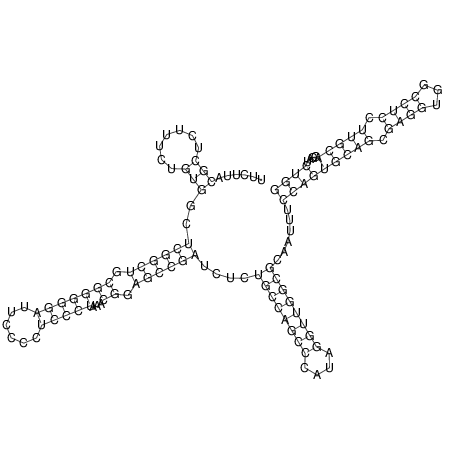
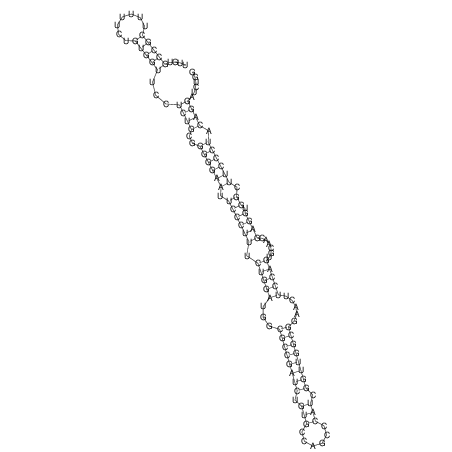
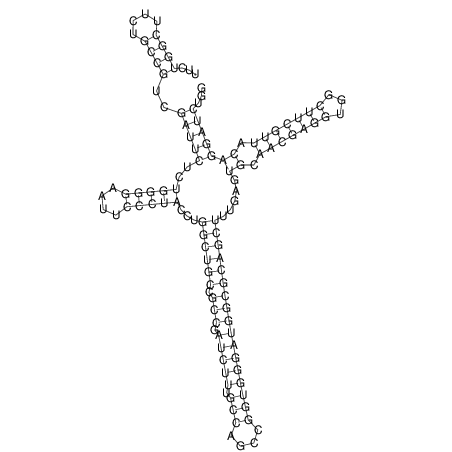
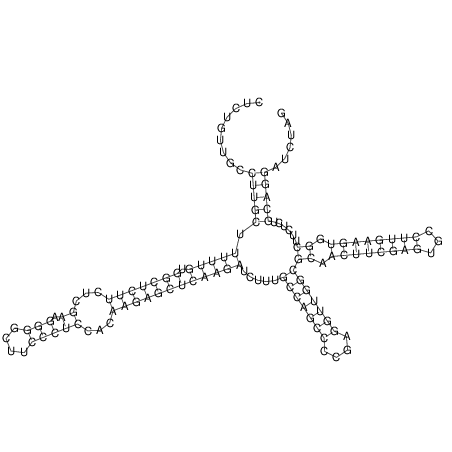


View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-4986 |
chr3L:9726758-9726880 - |
Known_mirbase |
|---|---|---|
| [View on miRBase] | [View on UCSC Genome Browser {Cornell Mirror}] | CDS |
Legend:
mature strand at position 9726828-9726850| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr3L:9726743-9726895 - | 1 | AGACTGCCAACTGGGTTCTGCC----GCTT---TTGCTGTGGCTTCTCTGCATGGG-ATTCCCCAT--TCTGCA-TGGCGCCGATCTCTGCCAGCCCATCGGATGGCGGAACTTCCAGTGCAGCGAGGTCGCTTCTCTGCAGGATCTGGTGGACTTGGGCG---------CCG | |
| droSim1 | chr3L:9107366-9107518 - | LASTZ | 5 | CAACTGCCAACTGGGTTCTGCC----GCTT---TTGCTGTGGCTTCTCTGCGTGGG-ATTCCCCAT--ACTGCA-TGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGATGGATTTGGGCG---------CCG |
| droSec1 | super_0:1960228-1960380 - | LASTZ | 5 | CAACTGCCAACTGGGTTCTGTC----GCTT---TTGCTGTGCCTTCTCTGCGTGGG-ATTCCCCAT--CCTGCA-TGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGCAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGGTGGACCTGGGCG---------CCG |
| droYak2 | chr3L:9698225-9698377 - | LASTZ | 4 | CAACTGCCAATTGGGTTCTGCC----GCTT---TTTCTGTGGCTTCTCTGCGCGGG-ATTCCCCAG--CCTGCA-TGGTGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGAATTGGGCG---------CCG |
| droEre2 | scaffold_4784:9713780-9713932 - | LASTZ | 4 | CAACTGCCAATTGGGTTCTGCC----GCTC---TTTCTGTGCCTTCTCTGCGCGGG-ATTCCCCAG--CCTGCA-TGGAGCTGATCTCTGCCAGCCCGTCGGGTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGACCTGGGCG---------CCG |
| droEug1 | scf7180000409711:1583067-1583219 - | LASTZ | 4 | TGACTGCCAAGTGGGTTCTGCC----GCTT---TTGCTATGGCTCTCTCTTGGGGG-ATTTCCCTT--GTTAAA-CGGCGTCGATCTCTGCCAGCCCATCGGTTGGCGGAATTTCCAGTGCAACGAGGTCGCATCTCTGCAGGATCTGGTAGATTTGGGAG---------CTG |
| droBia1 | scf7180000302428:8053551-8053703 + | LASTZ | 5 | CCGCTGCAAATAGGGTTCTGCC----ACTT---TTTCTGCTGCTTTTCTGCGGGGG-ATTCCCCGT--CCCGAA-CGCAGCCGATCTCTGCCAGCCCCTTGGCTGGCGGAACTTCCAGTGCAACGAGATCGCTTCCCTGCAGGATCTGGTGGACTTGGGGG---------CCG |
| droTak1 | scf7180000415741:49485-49637 + | LASTZ | 5 | TGACTGCCAATTGGGTTCTGCC----GCTT---TTTCTGTGGCTCTTCTATGGGGG-ATTTCCCCA--TGTGAG-CGGCGCCGATCTCTGCGAGCCCCTTGGATGGCGGAATTACCAGTGCAATGAGATCGCTTCCCTGCAGGATCTGGTGGATTTGGGAG---------CGG |
| droEle1 | scf7180000490564:510103-510255 - | LASTZ | 5 | TGACTGCCAATTGGTTTCTGCC----GCTT---TTTCTGTGGCTCGGCTGCGGGGG-ATTCCCCTC--GCTGAA-CGGAGCCGATCTCTGCCAGCCCGTTGGTTGGCGCAACTTCCAGTGCAACGAGGTGGCCTCTCTGCAGGATCTGGTGGATTTGGGAG---------CCG |
| droRho1 | scf7180000778255:38347-38499 - | LASTZ | 5 | TTTCTGCCAATTGGTTTCTTAC----GCTC---TTTCTGTGGCTCGGCTGCGGGGG-ATTCCCCTC--CCTAAA-CGGAGCCGATCTCTGCCAGCCCATAGGTTGGCGCAATTTCCAGTGCAGCGAGGTGGCCTCCTTGCAGGATCTGGTGGATCTAGGAG---------CCG |
| droFic1 | scf7180000453807:871186-871338 - | LASTZ | 5 | CCACTGCCAATTGGGTTGTGCC----GCTT---TTTCTGTGGTTCCTCTGCGGGGG-AATTCCCTT--TCTGGA-TGGCGCCGATCTGTGCCAGCCCATCGGTTGGCGGAACTTCCAGTGCAACGAGGTGGCTTCCCTACAGGATCTGGTGGATTTGGGGG---------CAG |
| droKik1 | scf7180000302486:2298206-2298349 - | LASTZ | 5 | CAACTGCCGG----GTTCTG------GCTT---CTGCCGTCGATTCTCTGG---GG-AATTCCCTA--CCTGGC-TGCCGCCGATCTTTGCCAGCCGGTGGGATGGCGCAGCTTTGAGTGCAACGAGGTGGCTTCGTTACAGGATCTGGTGGGTTTGGGGG---------CAG |
| droAna3 | scaffold_13337:3652327-3652482 + | LASTZ | 5 | TGCCAGTCAAATGGACTCTGTT----GCCTTGCTTTTTGTGGCTCTTCTCGAAGGG-GCTTCCCTC--CACAAG-AGCTCAAGATCTTTGCCAGCCCCGAGGTTGGCGCAACTTCGAGTGCCTTGAAGTGGCTTCTCTGCAGGATCTAGTGGATTTGGGGG---------CCG |
| droBip1 | scf7180000395954:52966-53124 + | LASTZ | 5 | TGACAGTCAAATGGATTCTGTT----GGCTGGCTTTTTGTGGCTTTTCCCGAAGGGGCTTTTTCCCGGCACAAG-AGCTCAAGATCTTTGCCAGCCCCGGGGTTGGCGCAATTTCGAGTGCCTTGAAGTGGCCTCTCTGGAGGATCTAGTGGATCTGGGGG---------CCG |
| dp4 | chrXR_group6:9932877-9933023 + | LASTZ | 5 | TGAATGCCAATTGGGTTTTGTC----CG------CTCTGTGGCTCTGCTGCAGGGG-CTTCCCCCA-----GGT-CGGGGCTCTTCCCTGCCGGACTCTGGCCTGGCGCAGCTTCGAGTGTCGTGAATTAGCCAGTCTGCAGGATCTGTCTGATGTGGGGG---------CCG |
| droPer1 | super_61:212952-213098 + | LASTZ | 5 | TGAATGCCAATTGGGTTTTGTC----CG------CTCTGTGGCTCTGCTGCAGGGG-CTTCCCCCA-----GGT-CGGGGCTCTTCCCTGCCGGACTCTGGCCTGGCGCAGCTTCGAGTGTCGTGAATTAGCCAGTCTGCAGGATCTGTCTGATGTGGGGG---------CCG |
| droWil1 | scaffold_180949:1195301-1195459 + | LASTZ | 5 | CGAATTTCCAGTGGTTTTGGCC----GTTG---GCTTTATGGCTTGTATGTCGGTG-CTTCCCGC---GGTGCA-CA--GCTCAAGTCTGTAATCCCCGCGGCTGGCGTAACTACGAATGCAGCGAATTGGCTAGTCTAGAGGATTTAATAAATTCTGAAGGCTTGTCCACTG |
| droVir3 | scaffold_13049:8736379-8736540 + | LASTZ | 5 | CGAATGCCAAACAG-TTCTGGACTTGGTCA---GTTCTCTGGCTGCTCTGCTGGGC-TGTGCCCCG--TG--CGTTG--GCGCAGTTGTGCGAGGCGCAGGGCTGGCGCAGCTTTGAGTGCCGCGAGCTGGGCAGCCTGCAGGAGCTGATCAATGAGAAGGCCTTGCCCACTG |
| droMoj3 | scaffold_6680:6608267-6608428 - | LASTZ | 5 | TGAATGCGCAACCT-TTGCGCCTAGGTCCA---GTCCTCTGGCTGCTGTGCTGCGG-CCTTTCCCA--GGTG-C-TG--AGTCAGATATGCGAGGCTCAAGGCTGGCGCAGCTTCGAGTGTCGTGAGCTGAGCAGCCTGAAGGAGCTGAAGAACGTTAGGAACTTTCCCAAAG |
| droGri2 | scaffold_15110:4482444-4482593 + | LASTZ | 5 | TGTTCTACACTTGG-------------CCA---ATTCTTTGCCTGCTCTGCTTTGG-GTTTTCCC----CTGGATTG--GCTGTGGTCTGTGAGGCTCAAGGCTGGCGTAGTTTTGAGTGCCGCGAATTGAGCAGCCTGCAGGAGCTGATCGATGTGAAGGCATTTCCCACCG |
| Species | Alignment |
| dm3 | AGACTGCCAACTGGGTTCTGCC----GCTT---TTGCTGTGGCTTCTCTGCATGGG-ATTCCCCAT--TCTGCA-TGGCGCCGATCTCTGCCAGCCCATCGGATGGCGGAACTTCCAGTGCAGCGAGGTCGCTTCTCTGCAGGATCTGGTGGACTTGGGCG---------CCG |
| droSim1 | CAACTGCCAACTGGGTTCTGCC----GCTT---TTGCTGTGGCTTCTCTGCGTGGG-ATTCCCCAT--ACTGCA-TGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGATGGATTTGGGCG---------CCG |
| droSec1 | CAACTGCCAACTGGGTTCTGTC----GCTT---TTGCTGTGCCTTCTCTGCGTGGG-ATTCCCCAT--CCTGCA-TGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGCAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGGTGGACCTGGGCG---------CCG |
| droYak2 | CAACTGCCAATTGGGTTCTGCC----GCTT---TTTCTGTGGCTTCTCTGCGCGGG-ATTCCCCAG--CCTGCA-TGGTGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGAATTGGGCG---------CCG |
| droEre2 | CAACTGCCAATTGGGTTCTGCC----GCTC---TTTCTGTGCCTTCTCTGCGCGGG-ATTCCCCAG--CCTGCA-TGGAGCTGATCTCTGCCAGCCCGTCGGGTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGACCTGGGCG---------CCG |
| droEug1 | TGACTGCCAAGTGGGTTCTGCC----GCTT---TTGCTATGGCTCTCTCTTGGGGG-ATTTCCCTT--GTTAAA-CGGCGTCGATCTCTGCCAGCCCATCGGTTGGCGGAATTTCCAGTGCAACGAGGTCGCATCTCTGCAGGATCTGGTAGATTTGGGAG---------CTG |
| droBia1 | CCGCTGCAAATAGGGTTCTGCC----ACTT---TTTCTGCTGCTTTTCTGCGGGGG-ATTCCCCGT--CCCGAA-CGCAGCCGATCTCTGCCAGCCCCTTGGCTGGCGGAACTTCCAGTGCAACGAGATCGCTTCCCTGCAGGATCTGGTGGACTTGGGGG---------CCG |
| droTak1 | TGACTGCCAATTGGGTTCTGCC----GCTT---TTTCTGTGGCTCTTCTATGGGGG-ATTTCCCCA--TGTGAG-CGGCGCCGATCTCTGCGAGCCCCTTGGATGGCGGAATTACCAGTGCAATGAGATCGCTTCCCTGCAGGATCTGGTGGATTTGGGAG---------CGG |
| droEle1 | TGACTGCCAATTGGTTTCTGCC----GCTT---TTTCTGTGGCTCGGCTGCGGGGG-ATTCCCCTC--GCTGAA-CGGAGCCGATCTCTGCCAGCCCGTTGGTTGGCGCAACTTCCAGTGCAACGAGGTGGCCTCTCTGCAGGATCTGGTGGATTTGGGAG---------CCG |
| droRho1 | TTTCTGCCAATTGGTTTCTTAC----GCTC---TTTCTGTGGCTCGGCTGCGGGGG-ATTCCCCTC--CCTAAA-CGGAGCCGATCTCTGCCAGCCCATAGGTTGGCGCAATTTCCAGTGCAGCGAGGTGGCCTCCTTGCAGGATCTGGTGGATCTAGGAG---------CCG |
| droFic1 | CCACTGCCAATTGGGTTGTGCC----GCTT---TTTCTGTGGTTCCTCTGCGGGGG-AATTCCCTT--TCTGGA-TGGCGCCGATCTGTGCCAGCCCATCGGTTGGCGGAACTTCCAGTGCAACGAGGTGGCTTCCCTACAGGATCTGGTGGATTTGGGGG---------CAG |
| droKik1 | CAACTGCCGG----GTTCTG------GCTT---CTGCCGTCGATTCTCTGG---GG-AATTCCCTA--CCTGGC-TGCCGCCGATCTTTGCCAGCCGGTGGGATGGCGCAGCTTTGAGTGCAACGAGGTGGCTTCGTTACAGGATCTGGTGGGTTTGGGGG---------CAG |
| droAna3 | TGCCAGTCAAATGGACTCTGTT----GCCTTGCTTTTTGTGGCTCTTCTCGAAGGG-GCTTCCCTC--CACAAG-AGCTCAAGATCTTTGCCAGCCCCGAGGTTGGCGCAACTTCGAGTGCCTTGAAGTGGCTTCTCTGCAGGATCTAGTGGATTTGGGGG---------CCG |
| droBip1 | TGACAGTCAAATGGATTCTGTT----GGCTGGCTTTTTGTGGCTTTTCCCGAAGGGGCTTTTTCCCGGCACAAG-AGCTCAAGATCTTTGCCAGCCCCGGGGTTGGCGCAATTTCGAGTGCCTTGAAGTGGCCTCTCTGGAGGATCTAGTGGATCTGGGGG---------CCG |
| dp4 | TGAATGCCAATTGGGTTTTGTC----CG------CTCTGTGGCTCTGCTGCAGGGG-CTTCCCCCA-----GGT-CGGGGCTCTTCCCTGCCGGACTCTGGCCTGGCGCAGCTTCGAGTGTCGTGAATTAGCCAGTCTGCAGGATCTGTCTGATGTGGGGG---------CCG |
| droPer1 | TGAATGCCAATTGGGTTTTGTC----CG------CTCTGTGGCTCTGCTGCAGGGG-CTTCCCCCA-----GGT-CGGGGCTCTTCCCTGCCGGACTCTGGCCTGGCGCAGCTTCGAGTGTCGTGAATTAGCCAGTCTGCAGGATCTGTCTGATGTGGGGG---------CCG |
| droWil1 | CGAATTTCCAGTGGTTTTGGCC----GTTG---GCTTTATGGCTTGTATGTCGGTG-CTTCCCGC---GGTGCA-CA--GCTCAAGTCTGTAATCCCCGCGGCTGGCGTAACTACGAATGCAGCGAATTGGCTAGTCTAGAGGATTTAATAAATTCTGAAGGCTTGTCCACTG |
| droVir3 | CGAATGCCAAACAG-TTCTGGACTTGGTCA---GTTCTCTGGCTGCTCTGCTGGGC-TGTGCCCCG--TG--CGTTG--GCGCAGTTGTGCGAGGCGCAGGGCTGGCGCAGCTTTGAGTGCCGCGAGCTGGGCAGCCTGCAGGAGCTGATCAATGAGAAGGCCTTGCCCACTG |
| droMoj3 | TGAATGCGCAACCT-TTGCGCCTAGGTCCA---GTCCTCTGGCTGCTGTGCTGCGG-CCTTTCCCA--GGTG-C-TG--AGTCAGATATGCGAGGCTCAAGGCTGGCGCAGCTTCGAGTGTCGTGAGCTGAGCAGCCTGAAGGAGCTGAAGAACGTTAGGAACTTTCCCAAAG |
| droGri2 | TGTTCTACACTTGG-------------CCA---ATTCTTTGCCTGCTCTGCTTTGG-GTTTTCCC----CTGGATTG--GCTGTGGTCTGTGAGGCTCAAGGCTGGCGTAGTTTTGAGTGCCGCGAATTGAGCAGCCTGCAGGAGCTGATCGATGTGAAGGCATTTCCCACCG |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
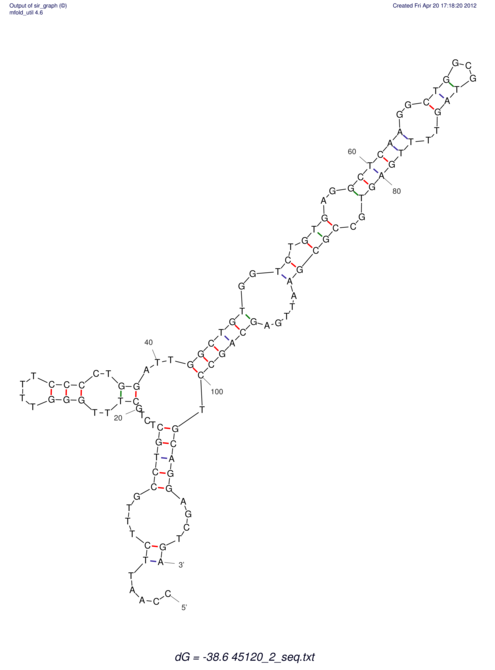
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:19:54 EDT