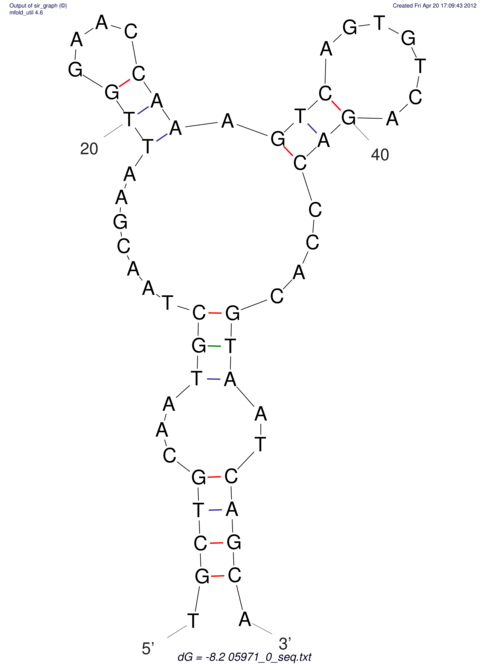
| dm3 |
chr2R:20181738-20181869 + |
NA |
ATAAAAAGTATGTCACT------------------TG-GGGG-------CGCTGCACT---------GGCATTGAAAAGT-GAATTACATTGATCGTGAC----AT-------GGGAATGGAAAATGTC-----G----CC-------------------------------------------------------------------------GAG---------------------------------------------------------------CCATGTAAATCGTTTGGCCTACCCGTCTACC-------A----GCCAACCAACC-------------------A |
| droSim1 |
chr2R:18643325-18643449 + |
1 |
ATAAAAAGTATGTCACT---------------TGGTG-GGGG-------CGCTGCACT---------GGCATTGAAAAGT-GAATTACATTGATCGTGAC----AT-------GGGGATGGAAAATGCC-----G----CC-------------------------------------------------------------------------GAG---------------------------------------------------------------CCATGTAAATCGATT----------------GAAGGCC-CG----TGGCCAACC-------------------A |
| droSec1 |
super_23:61187-61303 + |
3 |
ATAAAAAGTATGTCACT---------------TGGTG-GGGG-------CGCTGCACT---------GGCATTGAAAAGT-GAATTACATTGATCGTGAC----AT-------GGGGATGGAAAATGTC-----G----CC-------------------------------------------------------------------------GAG---------------------------------------------------------------CCATGTAAATCGATT----------------GAAGGCC-CG------------T-------------------G |
| droYak2 |
chr2R:20147877-20148015 + |
1 |
ATAAAAAGTATGTCACT------------------GG-GGGG-------CGCTGCACT---------GGCACTGAAAAGT-GAATTACATTGATCGTGAC----AT-------GGGGATGCGAAATGTC-----G----CCAAACGAAAAAA----TAAA------------------------------------------------------TCA---------------------------------------------------------------TAGTGTAACTCGTTT----------------GAAGGCCAC---ACCAACTAGTA-------------------A |
| droEre2 |
scaffold_4845:21546843-21546980 + |
10 |
ATAAAAAGTATGTCACT------------------AG-GGGG------CTTCTGCACT---------GGCATTGAAAAGT-GAATTACATTGATCGTGAC----AT-------GGGGATGGAAAATGTC-----G----CCGCACA-----------------------------------------------------------AAAAAA---GTG---------------------------------------------------------------TAGTGTAAATCGTTT----------------GAAGGCC-CATGGCCAACCAACC-------------------G |
| droEug1 |
scf7180000409464:540749-540858 + |
4 |
AAAAAAAGTATGTCACT------------------TG-GGGG-------CGCTGCACT---------GACACTGAAAAGT-GAATTACATTGATCATGGC----AG-------GGGAGCA--AAAT-------------------------------------------------------------CA---------------------------------------GGCA-------------------------------ATAG-------------CCATGTAAATCGTTA----------------GTA----------------AATC-------------------C |
| droBia1 |
scf7180000302143:729957-730086 + |
3 |
ACAAAAAGTATGTCACT------------------TG-GGGC-------CGCTGCACTGG-G----GGACATTGAAAAGT-GAATTACATTGATCATGGC----GT-------GGGGGC-----------------TGA---------A-AACTATCTA----------------------------------------GTAAATGGAAAA---GAG---------------------------------------------------------------TCGTGTAAACCGCTA-------------GT-G-AGTTC-CA--------------------------------T |
| droTak1 |
scf7180000415722:592281-592424 - |
3 |
CTAAAAAGTATGTCACT------------------TG-GGGG-------CGCTGCACTTGGG----GGACATTGAAAAGT-GAATTACATTGATCATGGC----ATATGGCATGGGGTCGG-------G-----TCTGA---------A-AACTGTCTA----------------------------------------GTAAATGGAAAA---GAG---------------------------------------------------------------TCTTGCAAATCGTTA----------------GTGAGC------------TCCAT-------------------G |
| droEle1 |
scf7180000491214:4867267-4867388 - |
4 |
ACAAAAAGTATGTCATT------------------TG-GGGG-------CGCTGCACT---------GGCATTGAAAAGT-GAATTACATTGATCATGGC----AT-------GGGGGCGCCAGAACTC-----A----CTGAAC----------------------------------------------------------------AA---GAG---------------------------------------------------------------CCATGTAAATCGTTA----------------GTGAGT-----------CCAATT-------------------C |
| droRho1 |
scf7180000777386:237447-237577 + |
2 |
AAAAAAAGTATGTCATT------------------TG-GGGG-------CGCTGCACT---------GACATTGAAAAGT-GAATTACATTGATCATGGC----AT-------GGGGGCGCCAAAACTC-----A----CCGAAC------------------------------------------------------------AGAAAA---GAG---------------------------------------------------------------TCATGTAAAACGTTTG---------------GTA----------------AATCCAATTCCAT----------G |
| droFic1 |
scf7180000453342:22911-23028 + |
1 |
A--AAAAGTATGTCAAA------------------TG-GGGG-------CGCCGAGGC---------GACATTGAAAAGT-GAATTACCCTGATCGTGGG----TG-------ACAGTCAGAAAAAAT-------------------------------------------------------------------------------AATA---GAG---------------------------------------------------------------CCATGTAAATGGTTA----------------GCA-GTT------------AGTAGTTCT--------------C |
| droKik1 |
scf7180000302277:1023196-1023338 - |
2 |
ATCAAAAGTATGTCAAC------------------TG-GGGGAGCGGAGTGCTGCACTGACC----TGACATTGAAAAGT-GAATTACAATGATCATGGC----GC---------------------------------------------------CAATAAATAGAGTGAGAC-----------------C---------------------GAG------------------------------AAGCCCGGTGGTGTCTT-G-------------GCCATGTAAATCGATG----------------G---------------GCATGCG-------------------T |
| droAna3 |
scaffold_13266:14229911-14230049 - |
5 |
AAAAAAAGTATGTCACA-----------------CTG-GGGG-------AACTGCACT---------GACATTGAAAAACTGAATTACATTGGACGGTGG----AT-------GGGT--GGA-----CTGATGGACTGATGGACT-------------------------GACAGCCACGA-----TCA--TCA---------------------------------------------------------------------CTAGCGTCGCAGTTAAGTTATGTAAACC--------------------------------------------------------------G |
| droBip1 |
scf7180000396759:1901653-1901780 + |
1 |
ATAAAAAGTATGTCACA-----------------CTG-GGGG-------AACTGCACT---------GACATTGAAAAATGGAATTACATTGAATGGTGG----AT-------GGGT--GGA-------------CT---------------------------------GACAGCCAAG--TTGATCAGATCA---------------------------------------------------------------------GTAGCTTTGTAGTTAAGTGATGTAAA-------------------------------------------CC-------------------G |
| dp4 |
chr3:18492568-18492741 - |
12 |
GTAAAAAGTATGTCAGAGAAGAGAAGAGAACACAGTGGGCGC-------TGCTGCACT---------GACATTGAAAAGT-GAATTGCATTGATCATGAC----GG-------GGGGTTGA-------C-----GCT---------------------------------GACTGCCACGAACCGCTCAGAACAATAGTGTAAATGTGGAATAATGC---------------------------------------------------------------TCGTAGAACCCATTC----------------GC-AGTC-CA-------TGGGCT-------------------G |
| droPer1 |
super_45:328754-328870 - |
26 |
GTGAAATGTATTTGAAA------------------CG------------------------------AGTAACGGAAATT-GCATTATTTCAATTACAAA----T----------------------C-----------------------------CAATGAGA----------------------------------------GGGAGA---GGG------ACAGGGACAGGGACCCAGC--------------------ACAAC----------------------------------CCTAGGAGCC-------A----GCCAGCCAGCC-------------------A |
| droWil1 |
scaffold_180949:4063354-4063494 - |
20 |
ATTGAGGGCGTGATTTA------------------TG-CCCG-------ATCTACCCC---------ATCATTTAATGTT-AA-AGACAATGCTAATTG-GCCAT---TAAAATAGGCCGCA---------------------------------------------------------------------------------------------ACAGACACAAATAGTTTGGGACCTAA------AAT-------------------------------------------CAAAGCCAACCGACCACC-------A----GCCAACCAGCC-------------------A |
| droVir3 |
scaffold_12875:7276052-7276211 + |
11 |
G---------------------------------------GG-------CGCTGCATTTGCCAGGCCAGAGTTAAAAAGT-GAATTGAATTGGCCGCTTG----AT-------TGAC--AG------CCGACGGGCTGA---------A-TA-TGTGCA----------------------------------------GCA-----------ATGC-------------CAACAAATCGATGCCAAAGT-----CAGTGTCAG-T-------------CCCACGTAA-TCGACA-------------ATC-------------------AGTTATTTA-CATACGAGCCACAA |
| droMoj3 |
scaffold_6473:1699609-1699741 - |
34 |
ATAATCTATTAGTTATT---------------TAATA-TATG-------T-----------------------------C-AA-TCATTCAGCCACTCA-ATCAT---TAAG---------------T-----------------------------TAATCAAC----------------------------------------G---------AGTAACTCAATCAGTCAGTCATTCAGT-----------------------GC-----------------CTCAATCGTTAAGTCTTCCAATAAGTC-------A----CTCAATCAGTC-------------------A |
| droGri2 |
scaffold_15203:4626832-4626941 - |
59 |
GCCCATGCCA--------------------------------------------------------------TAAAAAGA-GTTTTGAACTAATCTTAAG----CA-------GGCAGCGCAAATCGCT-----A----TAAATCAG-CTTT----CTA------------------------------------------------------------------------------------------------------------------------------TTCTTTGTTTTGCCCCACTGTGCACC-------A----GCCAGCCAGCC-------------------A |