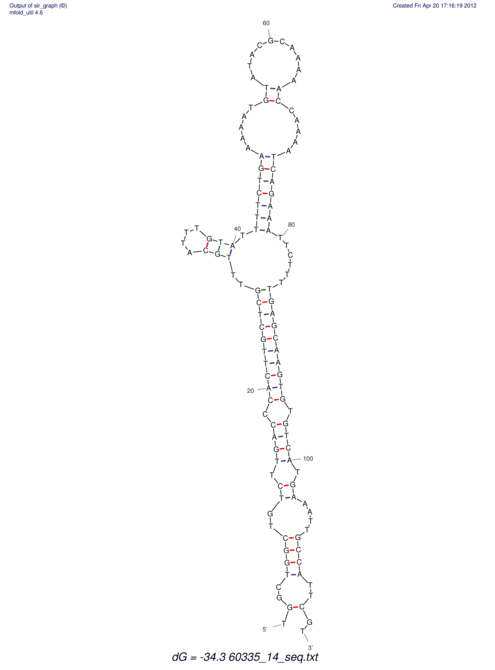
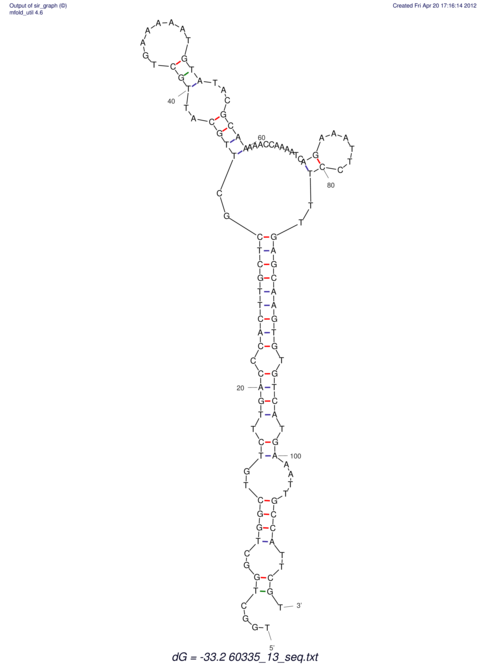
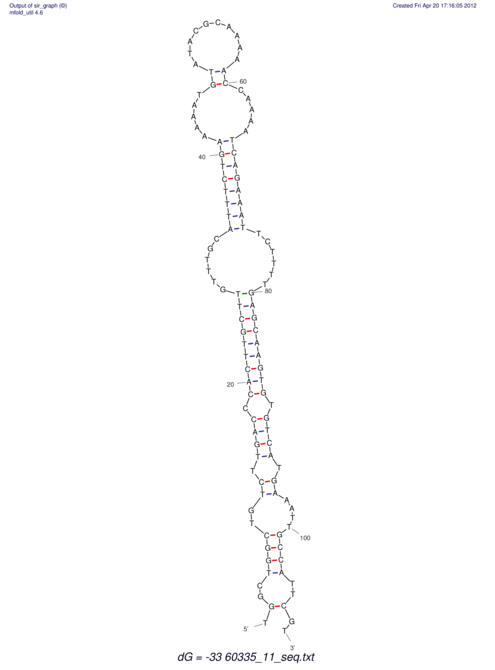
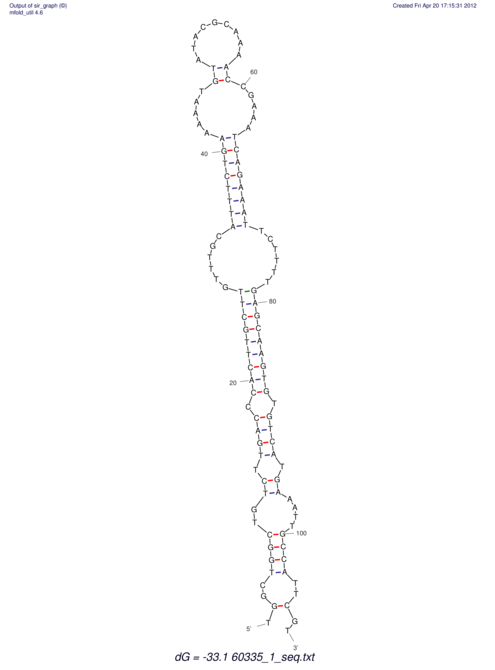
| dm3 |
chr2R:15524903-15525041 + |
|
1 |
A--ACA-CATCTGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-CG--TTTGCATTT-------CTG------AAAA---ATGTATACGCAA-AAA-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droSim1 |
chr2R:14228631-14228769 + |
LASTZ |
4 |
A--ACA-CATCTGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-CG--TTTGCATTT-------CTG------AAAA---ATGTATACGCAA-AAA-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droSec1 |
super_1:13039564-13039702 + |
LASTZ |
1 |
A--ACA-CATCTGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTG------AAAA---ATGTATACGCAA-AAA-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droYak2 |
chr2R:13649050-13649195 - |
LASTZ |
5 |
A--ACA-CATCTGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-CG--TTTGCATTTGTATTTTCTG------AAAA---ATGTATACGCAA-AAA-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droEre2 |
scaffold_4845:9723500-9723642 + |
LASTZ |
5 |
A--ACA-CATCTGCC---CGCTGG-CTGGCTG----------GCT----GT-------CTTGACCCACTTGCT-CG--CTTGCATTG-------CTG------AAAA---ATGTATACGCAA-AAA-CCAAAATCA------GAAATTCCTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droEug1 |
scf7180000409474:2425232-2425371 - |
LASTZ |
5 |
A--ACA-CATCTGCC--CCGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTG------AAAA---ATGTATACGCAA-AAA-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droBia1 |
scf7180000302143:3024933-3025070 + |
LASTZ |
5 |
A--ACA-CATCTGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTG------AAAA---ATGTATACGCAAA--A-CCGAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droTak1 |
scf7180000415384:129223-129366 - |
LASTZ |
5 |
A--ACA-CATCTGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTGAATCTGAAAA---ATGTATACGCAAA--A-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droEle1 |
scf7180000491201:1980229-1980378 + |
LASTZ |
5 |
A--ACA-CATCTGCC---CGCTGG-CTGGCTGGCTGGTTG--GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTG------AAAT---ATGTATACGCAAA--A-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droRho1 |
scf7180000780066:106697-106838 - |
LASTZ |
5 |
A--ACA-CATCTGCC---CGCTGG-CTGGCTG----------GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTG------AAAT---ATGTATACGCAAA--A-CCAAAATCA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droFic1 |
scf7180000454039:817245-817390 + |
LASTZ |
5 |
A--ACA-CATCTGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-CGCATTTGCATTT-------CTCAAAAAAAAAA---ATGTATACGCAAA--A-CCAAAATCC------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droKik1 |
scf7180000302679:61967-62124 + |
LASTZ |
5 |
A--ACAGCATCTGTGTGCTGCTGG-CTGG-----TGGCTGGCGCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTTCAAAAA--TG------AAAA---ATGTATACGCAA-AAACCCGAAAACA------GAAATTCTTTTGA-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATA---------------TAC |
| droAna3 |
scaffold_13266:9754397-9754555 - |
LASTZ |
5 |
AAAACA-CAAACGCC---CGCTG-------------------GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTCG-----CAAAAATATTCATATGCAAG--A-CCAAAATCAGAATCAGAAATTCTTTTGG-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCTCTATATGTA----CATCTAGTAC |
| droBip1 |
scf7180000396427:901727-901882 - |
LASTZ |
5 |
A--CCA-CAAACGCC---CGCTGG-CTG--------------GCT----GT-------CTTGACCCACTTGCT-TG--TTTGCATTT-------CTG------AAAAAATATTTATATGCAAG--A-CCAAAATCA------GAAATTCTTTTGG-GCAAGT----GTGTCATGAAATTGCC-ATTCGTTTCTGCTCT--ATATGTACATACATCTAGTAC |
| dp4 |
chr3:12652733-12652881 + |
LASTZ |
0 |
A--ACA-CGTCTCTC---CGTTTGTCAGTCTTTCAG------TCT----GTCAGCATGCTTGACCCACTTGATGTGCTTTTGAATTT-------TAG------AAAT---ATGTA---------AA-TCAAATTCT------GAAACATTTCGGGGACAAGTTTATGTGTCATTAAATTCCCCA----ATTTTATACT--AAA------------------ |
| droPer1 |
super_2:8929657-8929759 - |
LASTZ |
0 |
A--ACA-CTTCTCTC---CGTTTGTCAGTCTTTCAG------TCT----GTCAGCATGCTTGACCCACTTGATGTGCTTTTGAATTT-------TAG------AAAT---ATGTA---------AA-TCAAATTCT------GAAACATTT---------------------------------------------------------------------- |
| droWil1 |
Unknown |
|
0 |
----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_12875:4153222-4153295 - |
LASTZ |
0 |
A--ACA-CTTTTGAG---TGCCAG-CTT--------------TCTGAGCAT-------ATCAAA-CGCTCGA--CA--TTTGTCTTT-------TTG------AGGG---GGGTTCTCGCAC-A------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6496:20896701-20896727 + |
LASTZ |
0 |
A--ACA-CTTTTGAC---TGCTGC-CAG--------------CCT----GC-------C------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droGri2 |
Unknown |
|
0 |
----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |