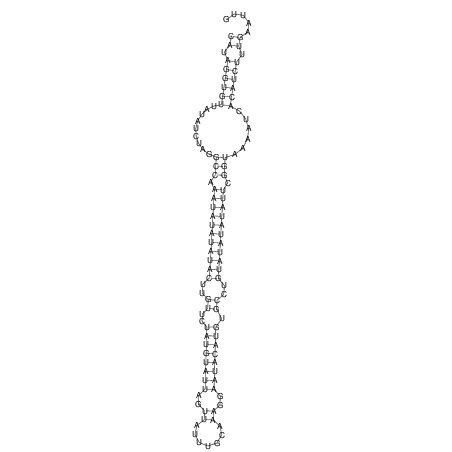
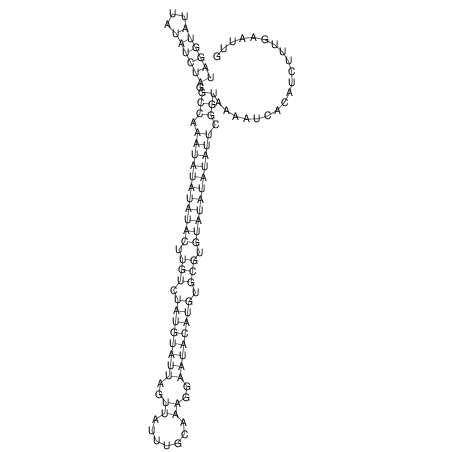
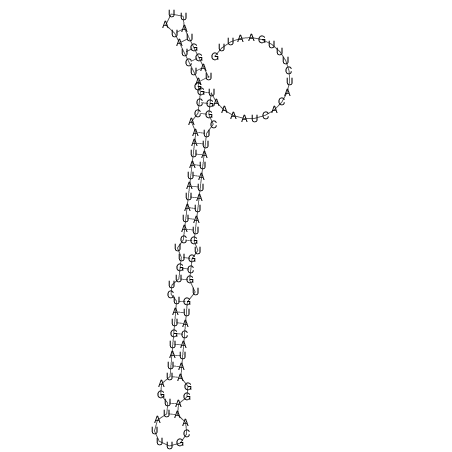
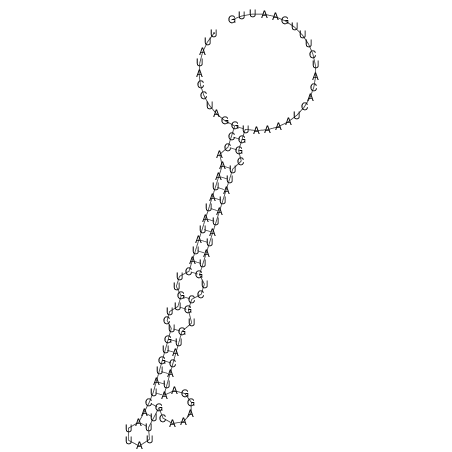
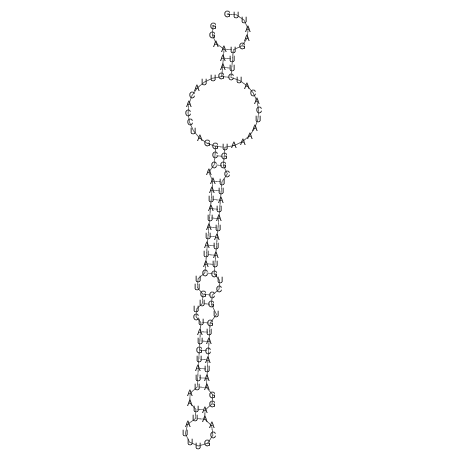
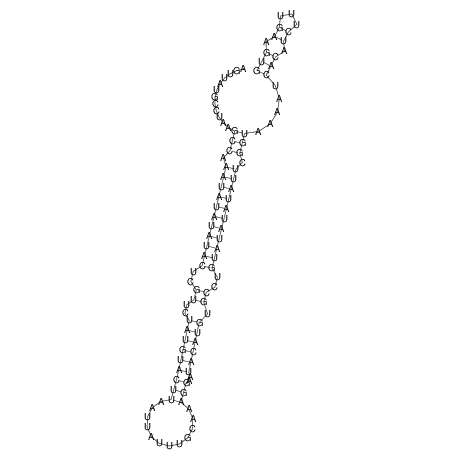
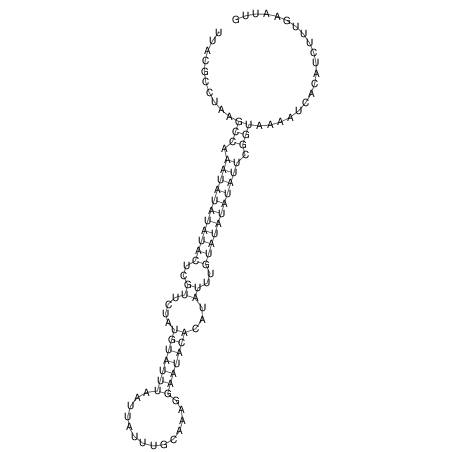
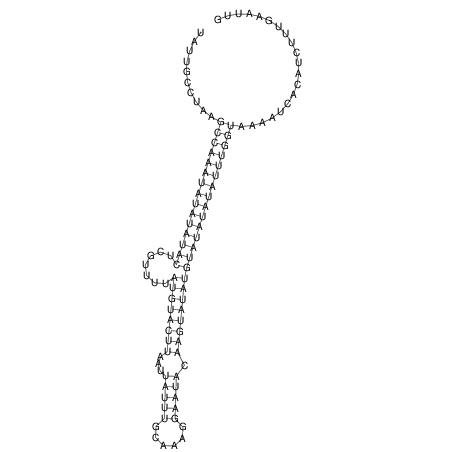

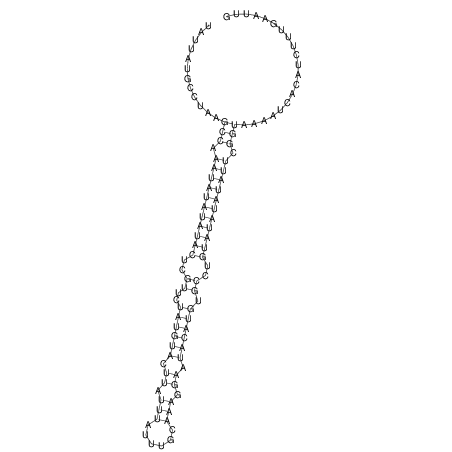
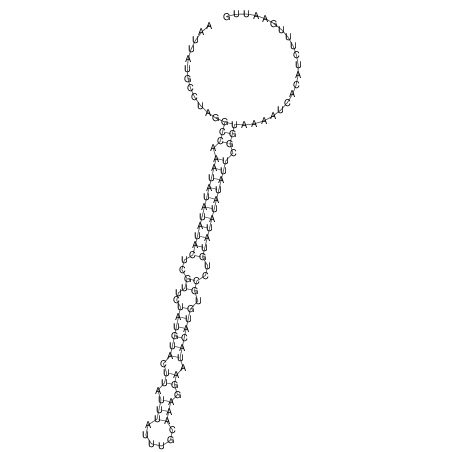
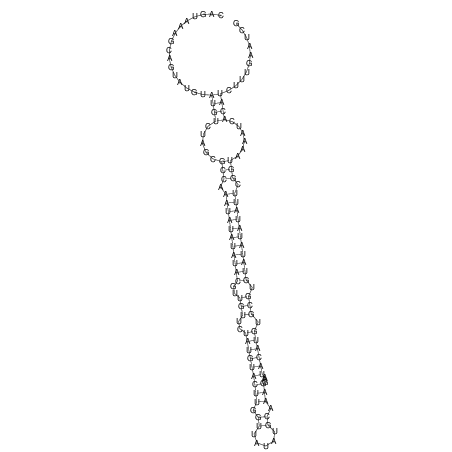
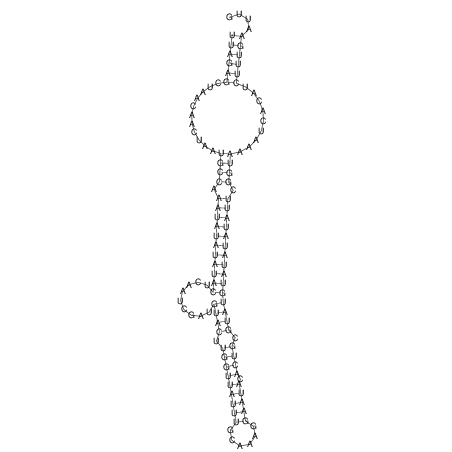
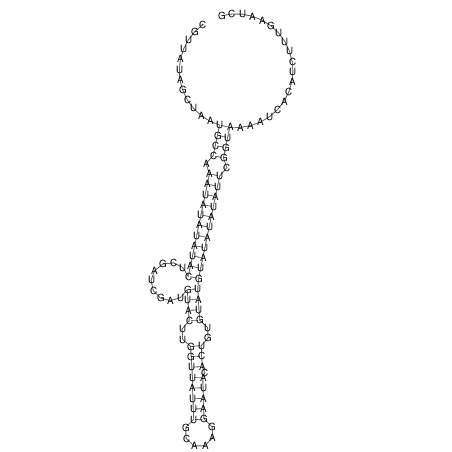

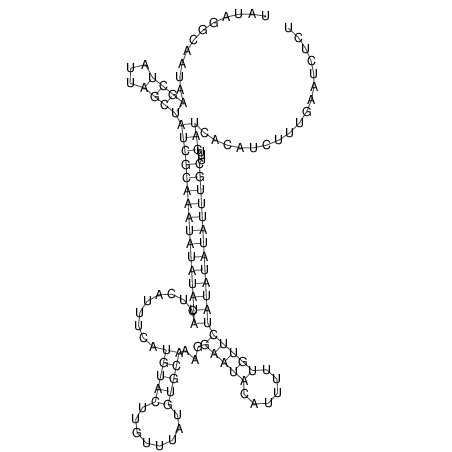
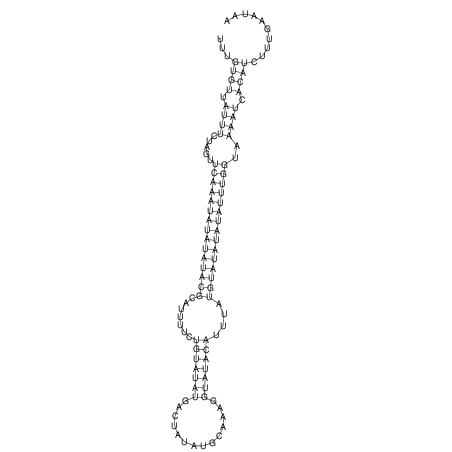
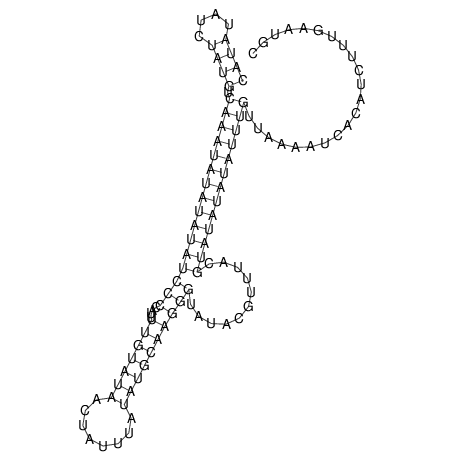
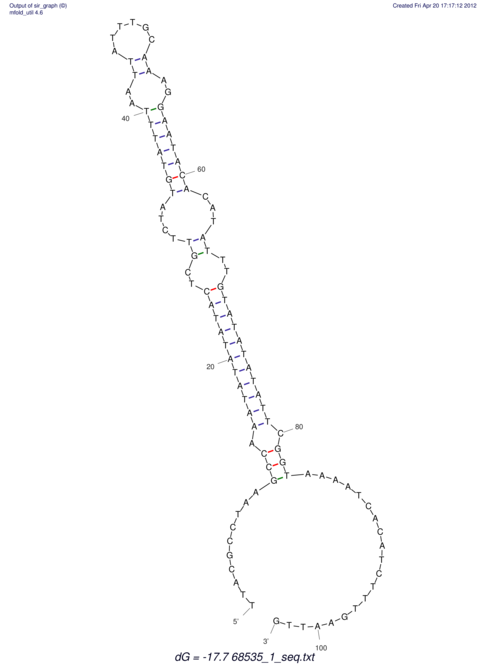
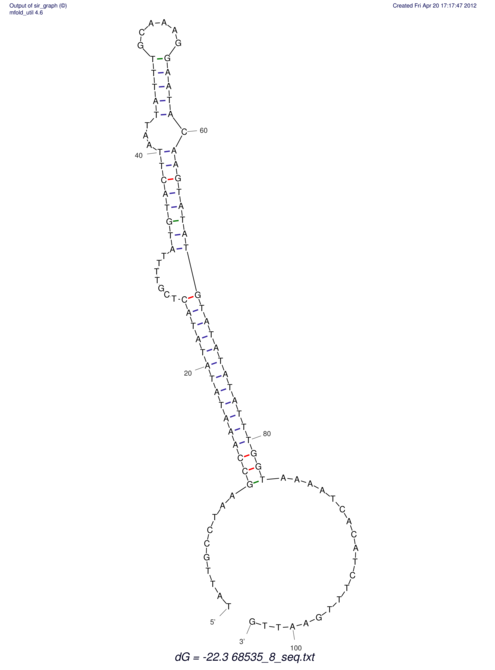
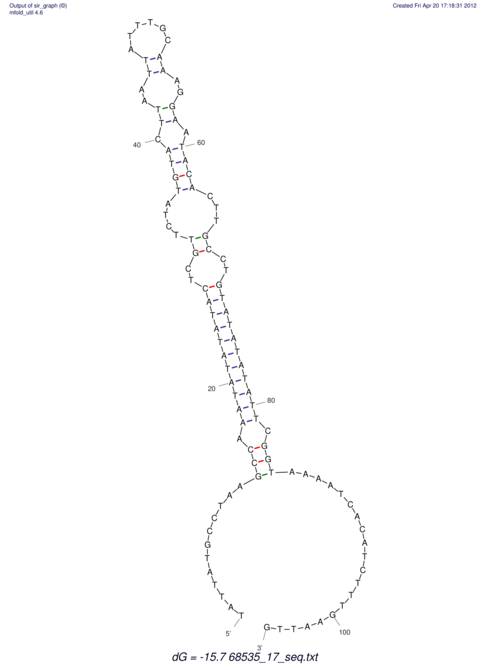
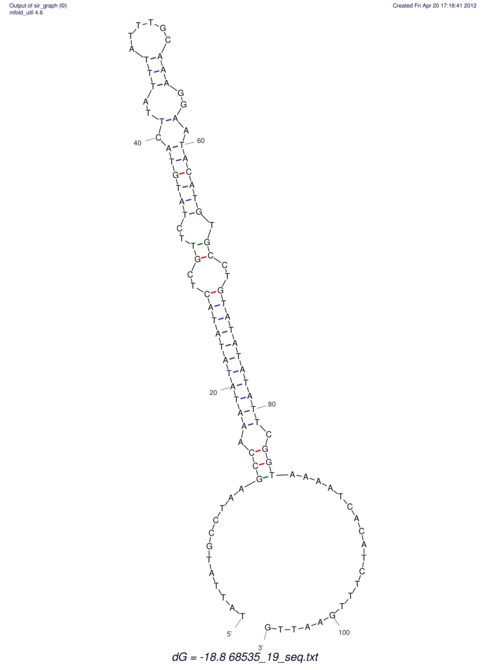

View alignment of MAF (12-files) vs LASTZ top hits (20-flies)
View all possible LASTZ hits (20-flies)
dme-mir-4979 |
chr2R:19621555-19621664 - |
Known_mirbase |
|---|---|---|
| [View on miRBase] | [View on UCSC Genome Browser {Cornell Mirror}] | intergenic |
Legend:
mature strand at position 19621580-19621601| Species | Coordinate | Source | Confidence | Alignment |
| dm3 | chr2R:19621540-19621679 - | 1 | GG-ATGGGAAA---------TTATACATAG---GT-----G-TTA-TATCTAG-GCCAAATATATATAC-TTGTTCTATGTA---TTAGTTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- | |
| droSim1 | chr2R:18187644-18187778 - | LASTZ | 1 | GG-ATGGGAAA---------CTA----TAG---GT-----A-TTA-TATCTAG-GCCAAATATATATAC-TTG-TCTATGTA---TTAGTTATTTGCAAAGGAATACAT-GTGCGTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droSec1 | super_9:2901906-2902041 - | LASTZ | 1 | GG-ATGGGAAA---------CTA----TAG---GT-----A-TTA-TATCTAG-GCCAAATATATATAC-TTGTTCTATGTA---TTAGTTATTTGCAAAGGAATACAT-GTGCGTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droYak2 | chr2R:19599592-19599718 - | LASTZ | 5 | TA-AGGGGAAA-------------------------------TTA-TACCTAG-GCCAAATATATATAC-TTGTTCTGTGTA---TCAATTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droEre2 | scaffold_4845:20984379-20984510 - | LASTZ | 5 | CT-GTAGAA------------------------GGAA-AAG-TTA-CACCTAG-GCCAAATATATATAC-TTGTTCTATGTA---TTAATTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droEug1 | scf7180000409474:21612-21738 + | LASTZ | 5 | CG----GGAA-----------------------------AG-TTA-TGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--CTTAATTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAG--TG-CCACAGCTCGCTAGT- |
| droBia1 | scf7180000302143:1773637-1773756 + | LASTZ | 5 | AG----------------------------------------TTA-CGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--TTTAATTATTTGCAAAGGAATACAC-ATATTTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACTGCTCGCTAGT- |
| droTak1 | scf7180000415400:558002-558129 - | LASTZ | 5 | AG-TTGGGAGA----------------------------TA-T---TGCCTAA-GCCAAATATATATAC-TCGTTTTATGTA--CTTAATTATTTGCAAAGGAATACAA-GTATATGTATATATATTTGGTAAAATCACATCTTTGAAT--TG-CCGGAGCTCGCTAGT- |
| droEle1 | scf7180000491201:1147757-1147882 + | LASTZ | 5 | GG------AAA----------------------------TA-TTA-TGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--CTTAATTATTTGCAAAGGAATACAC-TTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGTTAGT- |
| droRho1 | scf7180000778122:26363-26489 + | LASTZ | 5 | CA-----GAAA----------------------------TA-TTA-TGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--CTTATTTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droFic1 | scf7180000453809:586655-586785 + | LASTZ | 5 | AG-TTGGAAAA---------A------------------AA-TTA-TGCCTAG-GCCAAATATATATAC-TCGTTCTATGTA--CTTATTTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droKik1 | scf7180000302470:2102706-2102852 + | LASTZ | 5 | AA-AAT-GAAA---------TTC--CAGT-AAAGCAGT--ATGTA-TGTCTAGCGCCAAATATATATACGTTGTTCTATGTA--CTTGGTTATATGCAAAGGAATACAT-GTGCGTGTATATATATTCGGTAAAATCACATCTTTGAAT--CG-CCACAGCTCGTTAGTT |
| droAna3 | scaffold_13266:8260384-8260523 - | LASTZ | 5 | GG-TTCAATGA---------TTA----TTA---G-----AGCTAA-CAACTAATGCCAAATATATATAC-TCAATCGATGTA--CTTGGTTATTTGCAAAGGAATACACTGCGTATGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCAAAGCTTGTTAGT- |
| droBip1 | scf7180000396759:556673-556805 + | LASTZ | 5 | GT-TTTGCAAA---------T------------------CG-TTA-TAGCTAATGCCAAATATATATAC-TCGATCGATGTA--CTTGGTTATTTGCAAAGGAATACACTGTGTATGTATATATATTCGGTAAAATCACATCTTTGAAT--CG-CCATAGCTTGTTAGT- |
| dp4 | chr3:9376521-9376668 + | LASTZ | 5 | AC-G--AGAAA---------TGC--CATA-TAGGCAATAAA-CTATTAGCTAT-CGCAAATATATATAC-TCATTTCATGTA--CTTGTTTATGTGCAAAGGAATACATTTTTGTTCTATATATATTTGCTTTAATCACATCTTTGAATCTTT-CCATAGCTCGTTAGA- |
| droPer1 | super_4:4699742-4699885 + | LASTZ | 5 | GAGATGCCA------------------TA-TAGGCAATAAG-CTATTAGCTAT-CGCAAATATATATAC-TCATTTCATGTA--CTTGTTTATGTGCAAAGGAATACATTTTTGTTCTATATATATTTGCTTTGATCACATCTTTGAATCTCT-CCATAGCTCGTTAGA- |
| droWil1 | scaffold_181141:2403442-2403586 + | LASTZ | 5 | GT-GTAGGTATAAGTGTTGCCTT----TTT---GT-----G-TTA-TTTCTAG-TTCAAATATATATAC-GCATTTTCTGTA--TATGACTATATGCAAAGGTATACAT-TTA--TGTATATATATTTGGTAAAATCACATCTTTGAAT--AATCCATAGCTCGTCAGA- |
| droVir3 | scaffold_12823:519093-519222 + | LASTZ | 5 | GT-ATTAT------------CTA----C--------------ATA-TATCTAT-GTCAAATATATATAT-CCCCATTTTGTATAACTATTTATATGCAAGGGTATACGT-TTA--CGTATATATATTTGTTAAAATCACATCTTTGAAT--GCTATATAGCTCGTTAGC- |
| droMoj3 | scaffold_6496:318093-318227 + | LASTZ | 5 | AG-TTGCAGAA---------GTATTCATAT---GT-----A-CAA-AATCTTATATCAAATATATATAT-ATGTCCCAT----------TTGTATGCAAGGGTATGCGT-TTCTCTATATATATATTTGTTAAAATCACATCTTTGAAT--GGTATTTAGCTCGTTAGC- |
| droGri2 | scaffold_15245:6158007-6158151 - | MAF | 5 | TT-GCCAGGAG----------TC---TTA-CAACTAACT-ACTTA-TAACTAT-ATCAAATATATATAC-TCACTTTGTGTATAGCTACGTATAAGCAAAGGTATACGC-TTG--CGTATATATATTTGTTAAAATCACATCTTTGAAT--GGTATATAGCTCGTCAGC- |
| Species | Alignment |
| dm3 | GG-ATGGGAAA---------TTATACATAG---GT-----G-TTA-TATCTAG-GCCAAATATATATAC-TTGTTCTATGTA---TTAGTTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droSim1 | GG-ATGGGAAA---------CTA----TAG---GT-----A-TTA-TATCTAG-GCCAAATATATATAC-TTG-TCTATGTA---TTAGTTATTTGCAAAGGAATACAT-GTGCGTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droSec1 | GG-ATGGGAAA---------CTA----TAG---GT-----A-TTA-TATCTAG-GCCAAATATATATAC-TTGTTCTATGTA---TTAGTTATTTGCAAAGGAATACAT-GTGCGTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droYak2 | TA-AGGGGAAA-------------------------------TTA-TACCTAG-GCCAAATATATATAC-TTGTTCTGTGTA---TCAATTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droEre2 | CT-GTAGAA------------------------GGAA-AAG-TTA-CACCTAG-GCCAAATATATATAC-TTGTTCTATGTA---TTAATTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droEug1 | CG----GGAA-----------------------------AG-TTA-TGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--CTTAATTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAG--TG-CCACAGCTCGCTAGT- |
| droBia1 | AG----------------------------------------TTA-CGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--TTTAATTATTTGCAAAGGAATACAC-ATATTTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACTGCTCGCTAGT- |
| droTak1 | AG-TTGGGAGA----------------------------TA-T---TGCCTAA-GCCAAATATATATAC-TCGTTTTATGTA--CTTAATTATTTGCAAAGGAATACAA-GTATATGTATATATATTTGGTAAAATCACATCTTTGAAT--TG-CCGGAGCTCGCTAGT- |
| droEle1 | GG------AAA----------------------------TA-TTA-TGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--CTTAATTATTTGCAAAGGAATACAC-TTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGTTAGT- |
| droRho1 | CA-----GAAA----------------------------TA-TTA-TGCCTAA-GCCAAATATATATAC-TCGTTCTATGTA--CTTATTTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droFic1 | AG-TTGGAAAA---------A------------------AA-TTA-TGCCTAG-GCCAAATATATATAC-TCGTTCTATGTA--CTTATTTATTTGCAAAGGAATACAT-GTGCCTGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCACAGCTCGCTAGT- |
| droKik1 | AA-AAT-GAAA---------TTC--CAGT-AAAGCAGT--ATGTA-TGTCTAGCGCCAAATATATATACGTTGTTCTATGTA--CTTGGTTATATGCAAAGGAATACAT-GTGCGTGTATATATATTCGGTAAAATCACATCTTTGAAT--CG-CCACAGCTCGTTAGTT |
| droAna3 | GG-TTCAATGA---------TTA----TTA---G-----AGCTAA-CAACTAATGCCAAATATATATAC-TCAATCGATGTA--CTTGGTTATTTGCAAAGGAATACACTGCGTATGTATATATATTCGGTAAAATCACATCTTTGAAT--TG-CCAAAGCTTGTTAGT- |
| droBip1 | GT-TTTGCAAA---------T------------------CG-TTA-TAGCTAATGCCAAATATATATAC-TCGATCGATGTA--CTTGGTTATTTGCAAAGGAATACACTGTGTATGTATATATATTCGGTAAAATCACATCTTTGAAT--CG-CCATAGCTTGTTAGT- |
| dp4 | AC-G--AGAAA---------TGC--CATA-TAGGCAATAAA-CTATTAGCTAT-CGCAAATATATATAC-TCATTTCATGTA--CTTGTTTATGTGCAAAGGAATACATTTTTGTTCTATATATATTTGCTTTAATCACATCTTTGAATCTTT-CCATAGCTCGTTAGA- |
| droPer1 | GAGATGCCA------------------TA-TAGGCAATAAG-CTATTAGCTAT-CGCAAATATATATAC-TCATTTCATGTA--CTTGTTTATGTGCAAAGGAATACATTTTTGTTCTATATATATTTGCTTTGATCACATCTTTGAATCTCT-CCATAGCTCGTTAGA- |
| droWil1 | GT-GTAGGTATAAGTGTTGCCTT----TTT---GT-----G-TTA-TTTCTAG-TTCAAATATATATAC-GCATTTTCTGTA--TATGACTATATGCAAAGGTATACAT-TTA--TGTATATATATTTGGTAAAATCACATCTTTGAAT--AATCCATAGCTCGTCAGA- |
| droVir3 | GT-ATTAT------------CTA----C--------------ATA-TATCTAT-GTCAAATATATATAT-CCCCATTTTGTATAACTATTTATATGCAAGGGTATACGT-TTA--CGTATATATATTTGTTAAAATCACATCTTTGAAT--GCTATATAGCTCGTTAGC- |
| droMoj3 | AG-TTGCAGAA---------GTATTCATAT---GT-----A-CAA-AATCTTATATCAAATATATATAT-ATGTCCCAT----------TTGTATGCAAGGGTATGCGT-TTCTCTATATATATATTTGTTAAAATCACATCTTTGAAT--GGTATTTAGCTCGTTAGC- |
| droGri2 | TT-GCCAGGAG----------TC---TTA-CAACTAACT-ACTTA-TAACTAT-ATCAAATATATATAC-TCACTTTGTGTATAGCTACGTATAAGCAAAGGTATACGC-TTG--CGTATATATATTTGTTAAAATCACATCTTTGAAT--GGTATATAGCTCGTCAGC- |
| dm3 | droSim1 | droSec1 | droYak2 | droEre2 | droEug1 | droBia1 | droTak1 | droEle1 | droRho1 | droFic1 | droKik1 | droAna3 | droBip1 | dp4 | droPer1 | droWil1 | droVir3 | droMoj3 | droGri2 | |
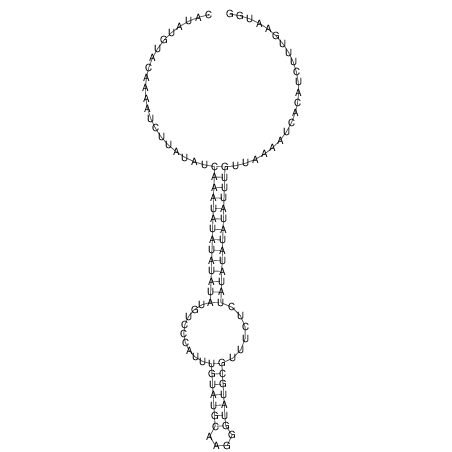
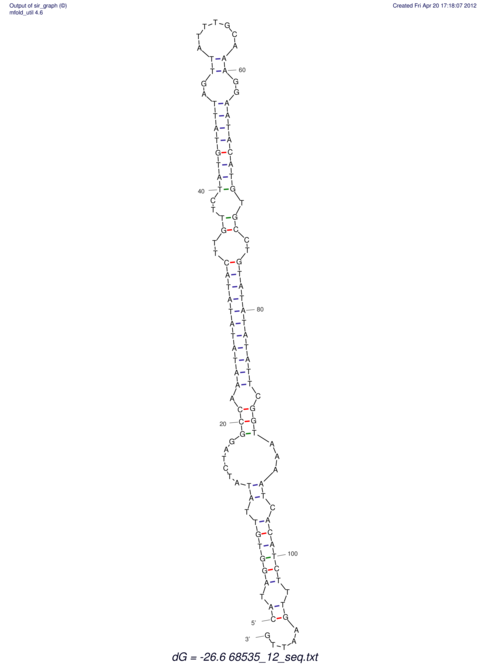
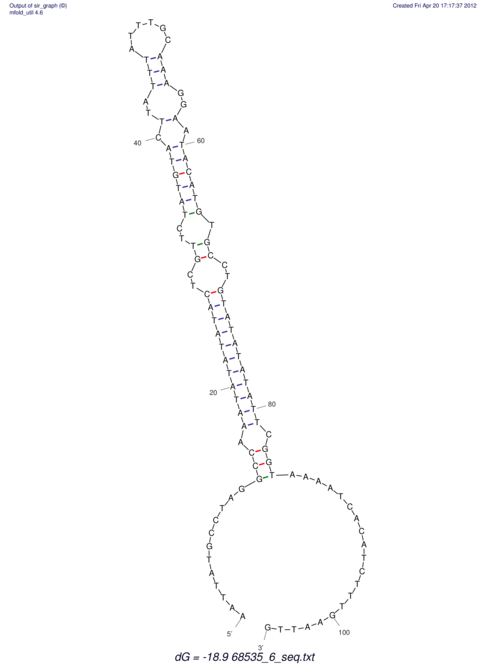
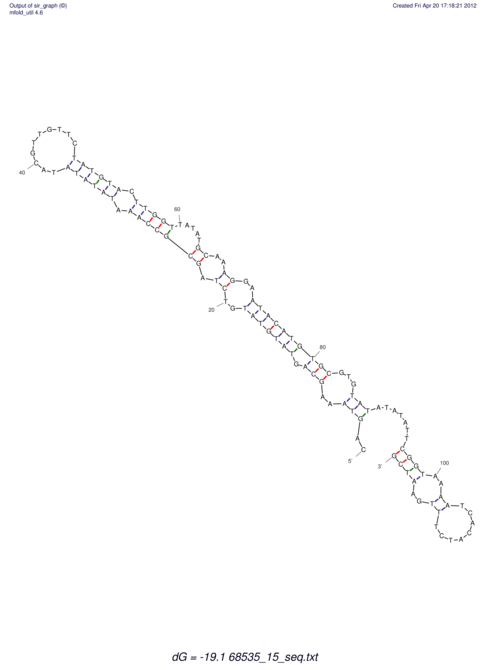
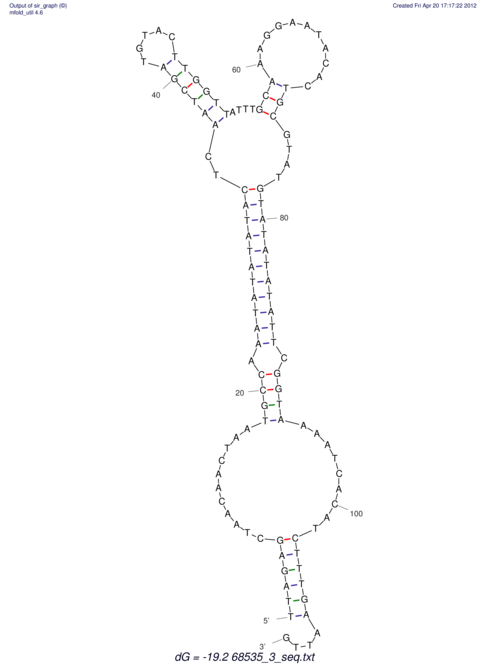
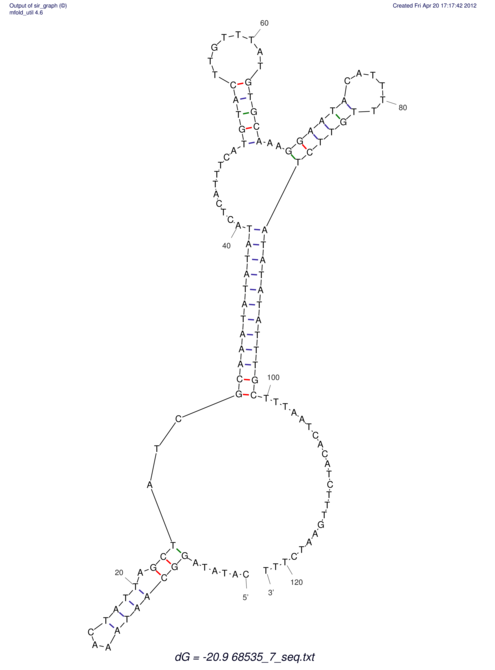
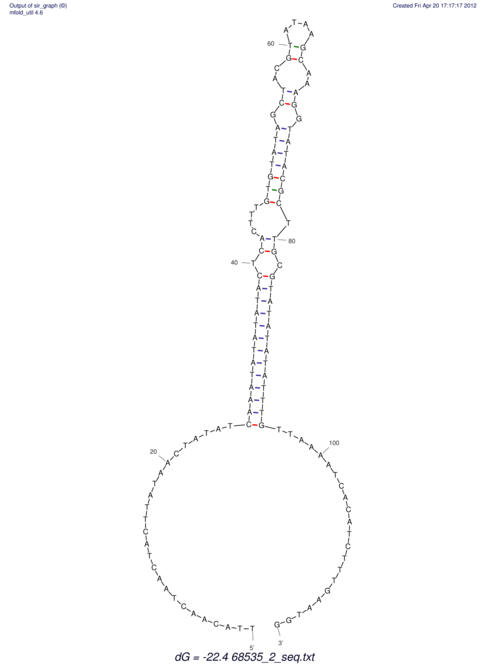
| RNAfold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
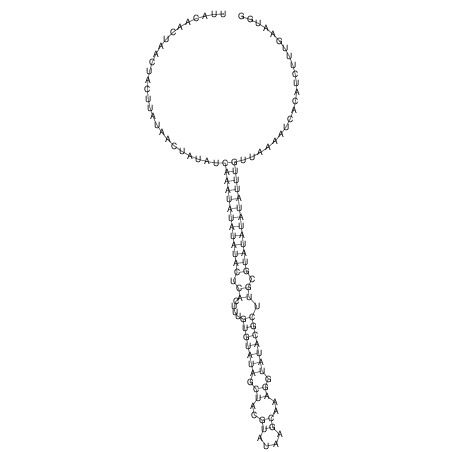
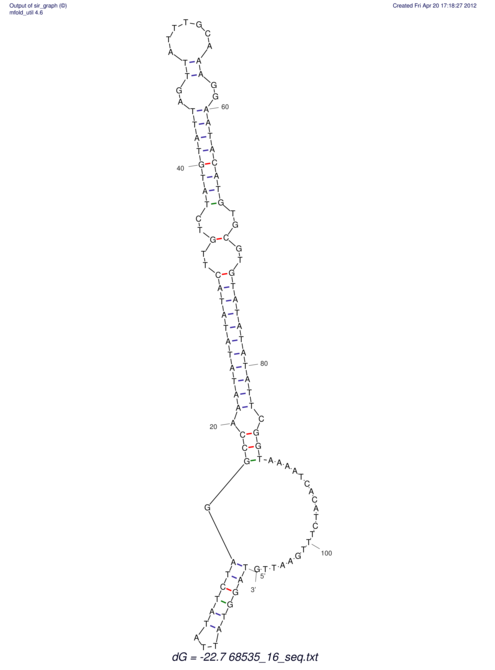
| UNAFold Image |  |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| Species | Alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 04/20/2012 at 05:18:45 EDT