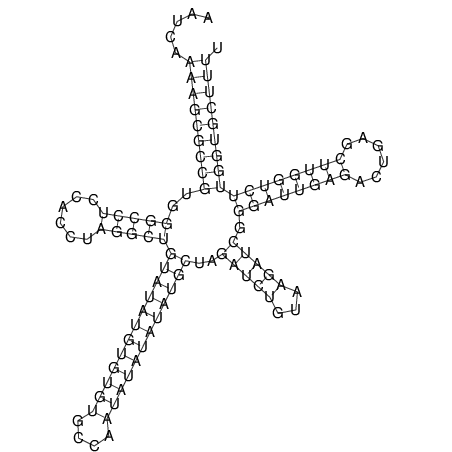
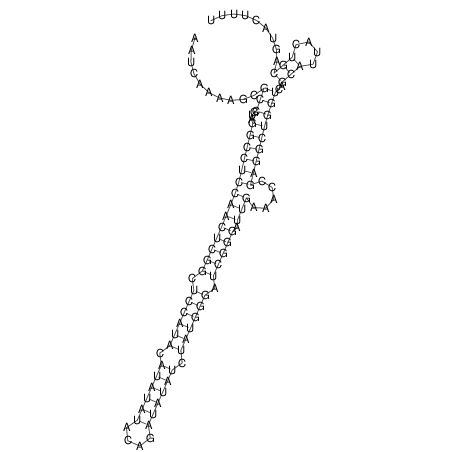
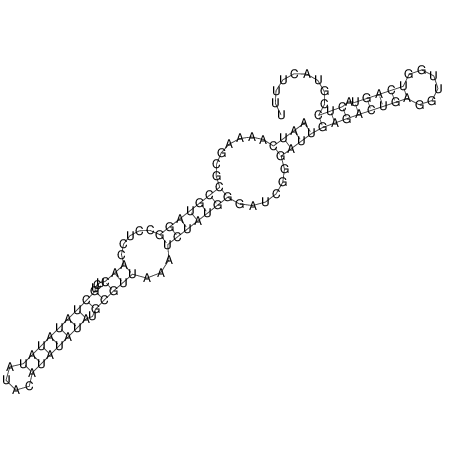
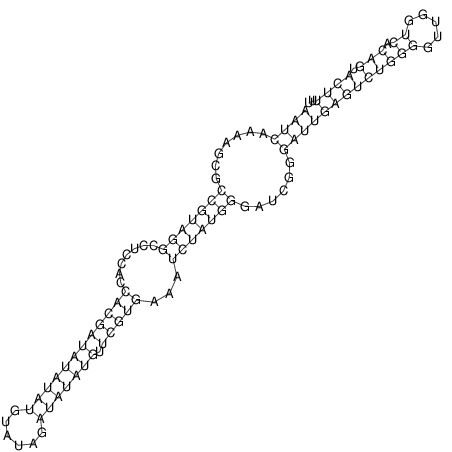
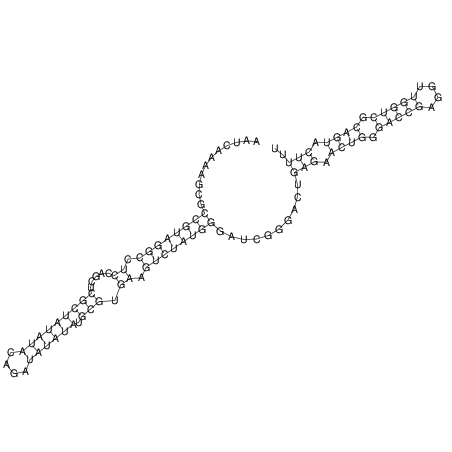
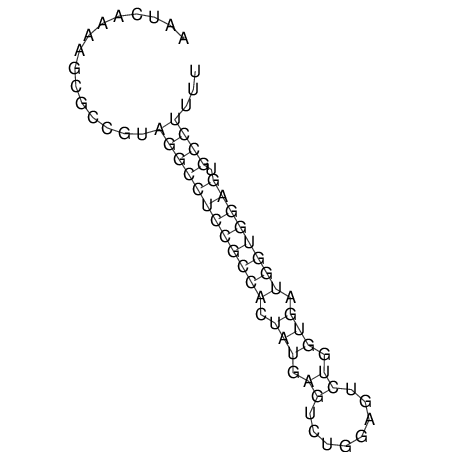
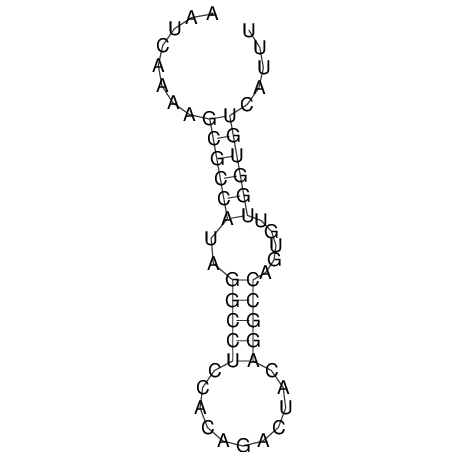
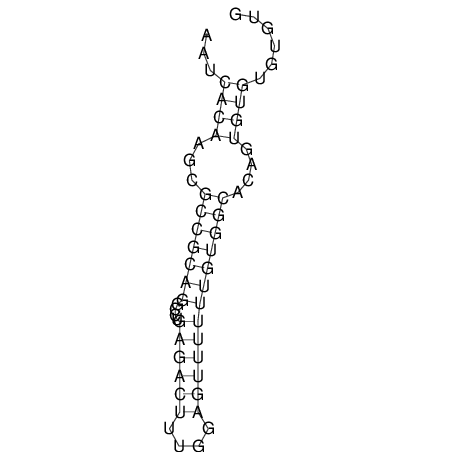
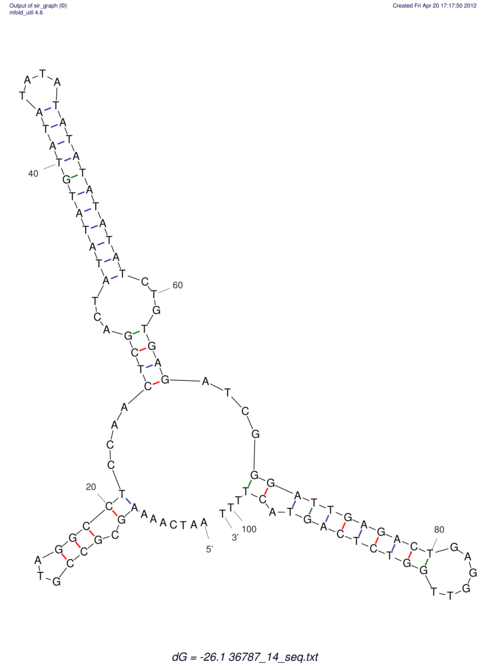
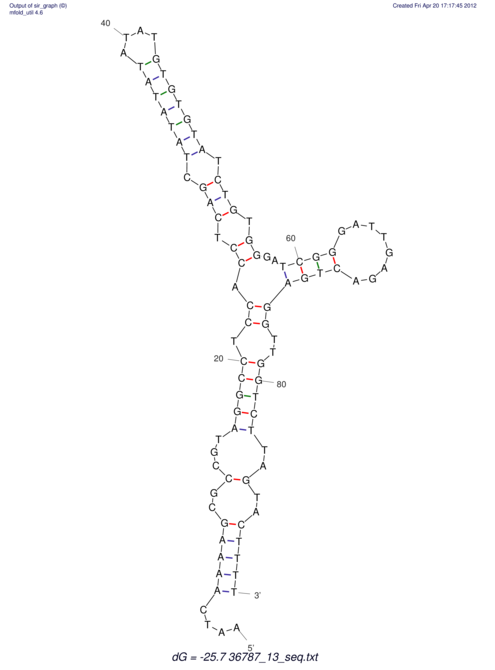
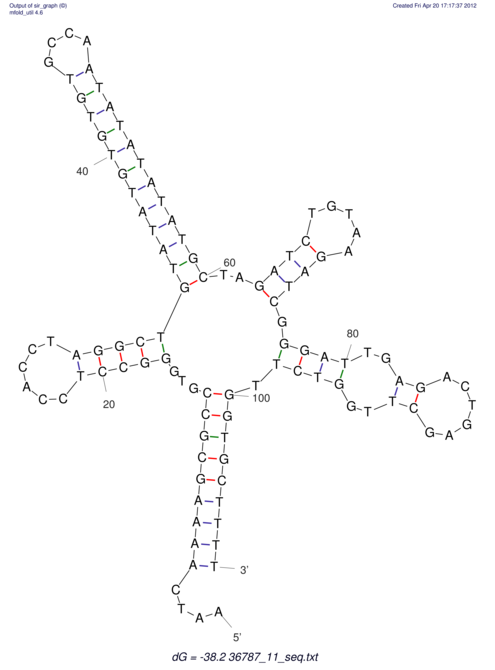
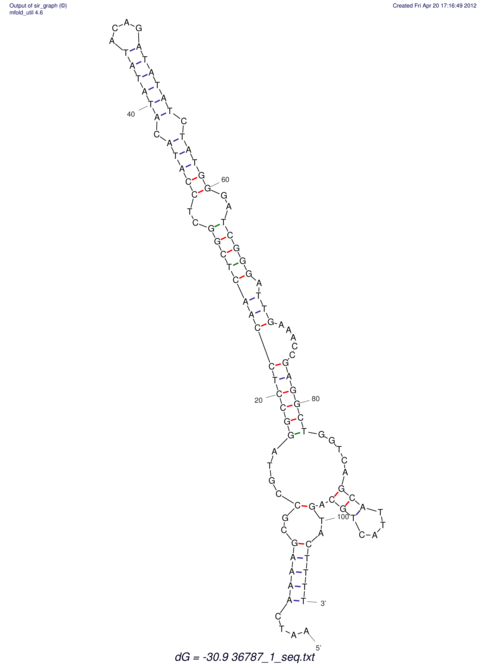
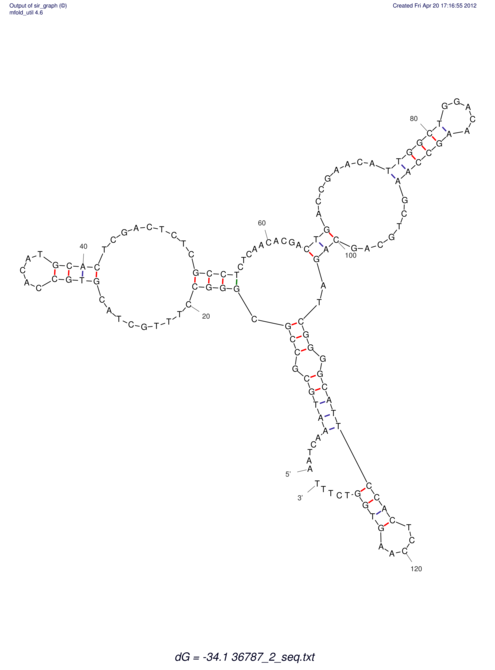
| CACTTGCGTGGCATAAATCACAAGCGCCGCAGGCCT-GAGACTTTG------GAGTT--------------TTTTGTGGCACAGTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | Size | Hit Count | Total Norm | V111
Male body | V042
Embryo | V050
Head | V057
Head | M021
Embryo |
| ...................................................................................GTATGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGT. | 18 | 62 | 0.05 | 0.02 | 0.00 | 0.00 | 0.00 | 0.03 |
| ...................................................................................GTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTTTT | 20 | 3 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTTTT | 19 | 6 | 0.50 | 0.33 | 0.00 | 0.00 | 0.17 | 0.00 |
| ...................................................................................GTGTGTAGATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGT. | 18 | 4 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 |
| ........................................................................GTTGTGGCACAGTGTATGT............................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ............................................................................CGGCGCAGTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G...... | 20 | 1 | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...............................................................................CGCAGTGTGTGTGTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT..... | 18 | 6 | 0.17 | 0.17 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..............................AGGACT-GAxACTTTG------GAGT.................................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| ..................................................................................AGTGTGTGTTTG----------------G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 20 | 4 | 0.25 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTACGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGT. | 18 | 8 | 0.13 | 0.00 | 0.13 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------xTGTGAG | 18 | 4 | 0.25 | 0.25 | 0.00 | 0.00 | 0.00 | 0.00 |
| ................................................................................ACAGTGTGTGTTTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGT... | 19 | 2 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTGTGTGTTTG----------------G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGT. | 18 | 47 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTGCGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 19 | 46 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTATGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTTT. | 18 | 4 | 0.25 | 0.00 | 0.00 | 0.00 | 0.25 | 0.00 |
| ...................................................................................GTATGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTATGTG | 19 | 8 | 0.13 | 0.13 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................................CGCGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGT. | 19 | 9 | 0.11 | 0.00 | 0.00 | 0.11 | 0.00 | 0.00 |
| ....................................................................................TGTGTGTTTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 21 | 6 | 0.17 | 0.17 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 19 | 3 | 0.33 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 |
| .................................................................................CTGTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTTT. | 20 | 5 | 0.20 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 20 | 42 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 |
| .....................................................................................GTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 19 | 49 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| ......................................................................................TGTGTTTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 19 | 7 | 0.14 | 0.14 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................................AGTGTGTGTGTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCGT. | 19 | 51 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 |
| ................................................................................ACTGTGTGTGCATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTG.. | 20 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................................TGTGTGTGTGTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 22 | 47 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 |
| ..................................................................................TGTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTATGT. | 19 | 45 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 |
| ..............................AGGACT-GAxACTTTG------GAGTT--------------................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................................TGTGTGTGTACG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGT. | 19 | 40 | 0.03 | 0.00 | 0.00 | 0.03 | 0.00 | 0.00 |
| ....................................................................................GATGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 18 | 11 | 0.09 | 0.09 | 0.00 | 0.00 | 0.00 | 0.00 |
| ..................................................................................AGTGTGTGTGTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGTGT. | 19 | 46 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................GTGTGTGACTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGT. | 18 | 46 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| .................................................................................TAGTGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTC.. | 19 | 2 | 0.50 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 |
| .....................................................................................GTGTGTGTG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 21 | 36 | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 |
| ...................................................................................ATGTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGTGTG | 20 | 47 | 0.02 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| .....................................................................................GTGTGTATG----------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGAGTG | 18 | 7 | 0.14 | 0.14 | 0.00 | 0.00 | 0.00 | 0.00 |